A set of reference sequences for the hepatitis C genotypes 4d, 4f, and 4k covering the full open reading frame
- PMID: 18551618
- PMCID: PMC2818806
- DOI: 10.1002/jmv.21240
A set of reference sequences for the hepatitis C genotypes 4d, 4f, and 4k covering the full open reading frame
Abstract
Infection with genotype 4 of the Hepatitis C virus is common in Africa and the Mediterranean area, but has also been found at increasing frequencies in injection drug users in Europe and North America. Full length viral sequences to characterize viral diversity and structure have recently become available mostly for subtype 4a, and studies in Egypt and Saudi Arabia, where high proportions of subtype 4a infected patients exist, have begun to establish optimized treatment regimens. However knowledge about other subtype variants of genotype 4 present in less developed African states is lacking. In this study the full coding region from so far poorly characterized variants of HCV genotype 4 was amplified and sequenced using a long range PCR technique. Sequences were analyzed with respect to phylogenetic relationship, possible recombination and prominent sequence characteristics compared to other known HCV strains. We present for the first time two full-length sequences from the HCV genotype 4k, in addition to five strains from HCV genotypes 4d and 4f. Reference sequences for accurate HCV genotyping are required for optimized treatment, and a better knowledge of the global viral sequence diversity is needed to guide vaccines or new drugs effective in the world wide epidemic.
Conflict of interest statement
The authors have no conflicting financial interests.
Figures

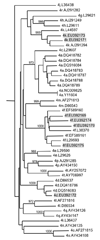



Similar articles
-
Full-length genome sequences of hepatitis C virus subtype 4f.J Gen Virol. 2007 Nov;88(Pt 11):2985-2990. doi: 10.1099/vir.0.83151-0. J Gen Virol. 2007. PMID: 17947520
-
Use of sequence analysis of the NS5B region for routine genotyping of hepatitis C virus with reference to C/E1 and 5' untranslated region sequences.J Clin Microbiol. 2007 Apr;45(4):1102-12. doi: 10.1128/JCM.02366-06. Epub 2007 Feb 7. J Clin Microbiol. 2007. PMID: 17287328 Free PMC article.
-
Sequence analysis of the core gene of 14 hepatitis C virus genotypes.Proc Natl Acad Sci U S A. 1994 Aug 16;91(17):8239-43. doi: 10.1073/pnas.91.17.8239. Proc Natl Acad Sci U S A. 1994. PMID: 8058787 Free PMC article.
-
Complete nucleotide sequence of genotype 4 hepatitis C viruses isolated from patients co-infected with human immunodeficiency virus type 1.Virus Res. 2007 Feb;123(2):161-9. doi: 10.1016/j.virusres.2006.09.001. Epub 2006 Oct 4. Virus Res. 2007. PMID: 17023084
-
Genetic variability of hepatitis C virus in South Egypt and its possible clinical implication.J Med Virol. 2009 Jun;81(6):1015-23. doi: 10.1002/jmv.21492. J Med Virol. 2009. PMID: 19382263
Cited by
-
Virological surveillance, molecular phylogeny, and evolutionary dynamics of hepatitis C virus subtypes 1a and 4a isolates in patients from Saudi Arabia.Saudi J Biol Sci. 2021 Mar;28(3):1664-1677. doi: 10.1016/j.sjbs.2020.11.089. Epub 2020 Dec 8. Saudi J Biol Sci. 2021. PMID: 33732052 Free PMC article.
-
Complete genomic sequences for hepatitis C virus subtypes 4b, 4c, 4d, 4g, 4k, 4l, 4m, 4n, 4o, 4p, 4q, 4r and 4t.J Gen Virol. 2009 Aug;90(Pt 8):1820-1826. doi: 10.1099/vir.0.010330-0. Epub 2009 Apr 8. J Gen Virol. 2009. PMID: 19357224 Free PMC article.
-
Expanded classification of hepatitis C virus into 7 genotypes and 67 subtypes: updated criteria and genotype assignment web resource.Hepatology. 2014 Jan;59(1):318-27. doi: 10.1002/hep.26744. Hepatology. 2014. PMID: 24115039 Free PMC article.
-
Highly Diverse Hepatitis C Strains Detected in Sub-Saharan Africa Have Unknown Susceptibility to Direct-Acting Antiviral Treatments.Hepatology. 2019 Apr;69(4):1426-1441. doi: 10.1002/hep.30342. Epub 2019 Mar 22. Hepatology. 2019. PMID: 30387174 Free PMC article.
-
Recombination in hepatitis C virus.Viruses. 2011 Oct;3(10):2006-24. doi: 10.3390/v3102006. Epub 2011 Oct 24. Viruses. 2011. PMID: 22069526 Free PMC article. Review.
References
-
- Airoldi A, Zavaglia C, Silini E, Tinelli C, Martinetti M, Asti M, Rossini A, Vangeli M, Salvaneschi L, Pinzello G. Lack of a strong association between HLA class II, tumour necrosis factor and transporter associated with antigen processing gene polymorphisms and virological response to alpha-interferon treatment in patients with chronic hepatitis C. Eur J Immunogenet. 2004;31(6):259–265. - PubMed
-
- Alfaleh FZ, Hadad Q, Khuroo MS, Aljumah A, Algamedi A, Alashgar H, Al-Ahdal MN, Mayet I, Khan MQ, Kessie G. Peginterferon alpha-2b plus ribavirin compared with interferon alpha-2b plus ribavirin for initial treatment of chronic hepatitis C in Saudi patients commonly infected with genotype 4. Liver Int. 2004;24(6):568–574. - PubMed
-
- Chamberlain RW, Adams N, Saeed AA, Simmonds P, Elliott RM. Complete nucleotide sequence of a type 4 hepatitis C virus variant, the predominant genotype in the Middle East. J Gen Virol. 1997;78(Pt 6):1341–1347. - PubMed
-
- Chinchai T, Labout J, Noppornpanth S, Theamboonlers A, Haagmans BL, Osterhaus AD, Poovorawan Y. Comparative study of different methods to genotype hepatitis C virus type 6 variants. J Virol Methods. 2003;109(2):195–201. - PubMed
-
- Chung RT, Monto A, Dienstag JL, Kaplan LM. Mutations in the NS5A region do not predict interferon-responsiveness in american patients infected with genotype 1b hepatitis C virus. J Med Virol. 1999;58(4):353–358. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

