Biochemical individuality reflected in chromatographic, electrophoretic and mass-spectrometric profiles
- PMID: 18551752
- PMCID: PMC2603028
- DOI: 10.1016/j.jchromb.2007.10.007
Biochemical individuality reflected in chromatographic, electrophoretic and mass-spectrometric profiles
Abstract
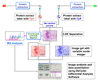
This review discusses the current trends in molecular profiling for the emerging systems biology applications. Historically, the methodological developments in separation science were coincident with the availability of new ionization techniques in mass spectrometry. Coupling miniaturized separation techniques with technologically-advanced MS instrumentation and the modern data processing capabilities are at the heart of current platforms for proteomics, glycomics and metabolomics. These are being featured here by the examples from quantitative proteomics, glycan mapping and metabolomic profiling of physiological fluids.
Figures

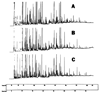


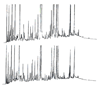


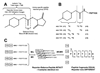
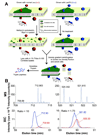

 , N-acetylglucosamine;
, N-acetylglucosamine;  , mannose;
, mannose;  , galactose;
, galactose;  , fucose;
, fucose;  , N-acetylneuraminic acid. Reproduced from [102] with permission.
, N-acetylneuraminic acid. Reproduced from [102] with permission.






Similar articles
-
Inductively coupled plasma mass spectrometric detection for chromatography and capillary electrophoresis.J Chromatogr A. 1997 Nov 21;789(1-2):85-126. doi: 10.1016/s0021-9673(97)00970-9. J Chromatogr A. 1997. PMID: 9440286 Review.
-
Applications of mass spectrometry in metabolomic studies of animal model and invertebrate systems.Brief Funct Genomic Proteomic. 2009 Jan;8(1):28-48. doi: 10.1093/bfgp/eln052. Epub 2008 Dec 12. Brief Funct Genomic Proteomic. 2009. PMID: 19074496 Review.
-
Elemental speciation by chromatographic separation with inductively coupled plasma mass spectrometry detection.J Chromatogr A. 2002 Oct 18;974(1-2):1-21. doi: 10.1016/s0021-9673(02)01239-6. J Chromatogr A. 2002. PMID: 12458926 Review.
-
[Applications of chromatography in glycomics].Se Pu. 2024 Jul;42(7):646-657. doi: 10.3724/SP.J.1123.2023.12003. Se Pu. 2024. PMID: 38966973 Free PMC article. Review. Chinese.
-
Recent liquid chromatographic-(tandem) mass spectrometric applications in proteomics.J Chromatogr A. 2003 Jun 6;1000(1-2):589-608. doi: 10.1016/s0021-9673(03)00178-x. J Chromatogr A. 2003. PMID: 12877191 Review.
Cited by
-
Metabolomics and malaria biology.Mol Biochem Parasitol. 2011 Feb;175(2):104-11. doi: 10.1016/j.molbiopara.2010.09.008. Epub 2010 Oct 21. Mol Biochem Parasitol. 2011. PMID: 20970461 Free PMC article. Review.
-
Omics as a Tool to Help Determine the Effectiveness of Supplements.Nutrients. 2022 Dec 14;14(24):5305. doi: 10.3390/nu14245305. Nutrients. 2022. PMID: 36558464 Free PMC article. Review.
-
Analysis of glycans derived from glycoconjugates by capillary electrophoresis-mass spectrometry.Electrophoresis. 2011 Dec;32(24):3467-81. doi: 10.1002/elps.201100342. Electrophoresis. 2011. PMID: 22180203 Free PMC article. Review.
References
-
- Kitano H. Science. 2002;295:1662. - PubMed
-
- van Ommen B, Stierum R. Curr. Opin. Biotechnol. 2002;13:517. - PubMed
-
- Morel MN, Holland JM, van der Greef J, Maple EW, Clish C, Loscalzo J, Naylor S. Mayo Clin. Proc. 2004;79:651. - PubMed
-
- van der Greef J, Stroobant P, van der Heijden R. Curr. Opin. Chem. Biol. 2004;8:559. - PubMed
-
- Williams RJ. Biochemical Individuality. New York: Wiley and Sons; 1956.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

