Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression
- PMID: 18552202
- PMCID: PMC2483357
- DOI: 10.1105/tpc.107.057380
Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression
Abstract
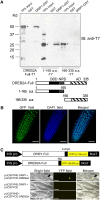
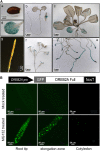
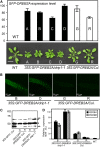
The DEHYDRATION-RESPONSIVE ELEMENT BINDING PROTEIN2A (DREB2A) transcription factor controls water deficit-inducible gene expression and requires posttranslational modification for its activation. The activation mechanism is not well understood; however, the stability of this protein in the nucleus was recently found to be important for its activation. Here, we report the isolation of Arabidopsis thaliana DREB2A-INTERACTING PROTEIN1 (DRIP1) and DRIP2, C3HC4 RING domain-containing proteins that interact with the DREB2A protein in the nucleus. An in vitro ubiquitination assay showed that they function as E3 ubiquitin ligases and are capable of mediating DREB2A ubiquitination. Overexpression of DRIP1 in Arabidopsis delayed the expression of DREB2A-regulated drought-responsive genes. Drought-inducible gene expression was slightly enhanced in the single T-DNA mutants of drip1-1 and drip2-1. By contrast, significantly enhanced gene expression was revealed in the drip1 drip2 double mutant under dehydration stress. Collectively, these data imply that DRIP1 and DRIP2 function negatively in the response of plants to drought stress. Moreover, overexpression of full-length DREB2A protein was more stable in drip1-1 than in the wild-type background. These results suggest that DRIP1 and DRIP2 act as novel negative regulators in drought-responsive gene expression by targeting DREB2A to 26S proteasome proteolysis.
Figures





Comment in
-
Ubiquitin ligation RINGs twice: redundant control of plant processes by E3 ubiquitin ligases.Plant Cell. 2008 Jun;20(6):1424. doi: 10.1105/tpc.108.200611. Epub 2008 Jun 13. Plant Cell. 2008. PMID: 18552197 Free PMC article. No abstract available.
References
-
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24 701–713. - PubMed
-
- Barlow, P.N., Luisi, B., Milner, A., Elliott, M., and Everett, R. (1994). Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy. A new structural class of zinc-finger. J. Mol. Biol. 237 201–211. - PubMed
-
- Bartels, D., and Sunkar, R. (2005). Drought and salt tolerance in plants. Crit. Rev. Plant Sci. 24 23–58.
-
- Borden, K.L. (2000). RING domains: Master builders of molecular scaffolds? J. Mol. Biol. 295 1103–1112. - PubMed
-
- Borden, K.L., and Freemont, P.S. (1996). The RING finger domain: A recent example of a sequence-structure family. Curr. Opin. Struct. Biol. 6 395–401. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

