Involvement of Saccharomyces cerevisiae Avo3p/Tsc11p in maintaining TOR complex 2 integrity and coupling to downstream signaling
- PMID: 18552287
- PMCID: PMC2519779
- DOI: 10.1128/EC.00065-08
Involvement of Saccharomyces cerevisiae Avo3p/Tsc11p in maintaining TOR complex 2 integrity and coupling to downstream signaling
Abstract
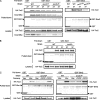
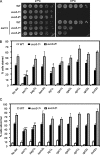
Target-of-rapamycin proteins (TORs) are Ser/Thr kinases serving a central role in cell growth control. TORs function in two conserved multiprotein complexes, TOR complex 1 (TORC1) and TORC2; the mechanisms underlying their actions and regulation are not fully elucidated. Saccharomyces TORC2, containing Tor2p, Avo1p, Avo2p, Avo3p/Tsc11p, Bit61p, and Lst8p, regulates cell integrity and actin organization. Two classes of avo3 temperature-sensitive (avo3(ts)) mutants that we previously identified display cell integrity and actin defects, yet one is suppressed by AVO1 while the other is suppressed by AVO2 or SLM1, defining two TORC2 downstream signaling mechanisms, one mediated by Avo1p and the other by Avo2p/Slm1p. Employing these mutants, we explored Avo3p functions in TORC2 structure and signaling. By observing binary protein interactions using coimmunoprecipitation, we discovered that the composition of TORC2 and its recruitment of the downstream effectors Slm1p and Slm2p were differentially affected in different avo3(ts) mutants. These molecular defects can be corrected only by expressing AVO3, not by expressing suppressors, highlighting the role of Avo3p as a structural and signaling scaffold for TORC2. Phenotypic modifications of avo3(ts) mutants by deletion of individual Rho1p-GTPase-activating proteins indicate that two TORC2 downstream signaling branches converge on Rho1p activation. Our results also suggest that Avo2p/Slm1p-mediated signaling, but not Avo1p-mediated signaling, links to Rho1p activation specifically through the Rho1p-guanine nucleotide exchange factor Tus1p.
Figures







Similar articles
-
Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-target-of-rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae.J Biol Chem. 2012 Feb 24;287(9):6089-99. doi: 10.1074/jbc.M111.303701. Epub 2011 Dec 28. J Biol Chem. 2012. PMID: 22207764 Free PMC article.
-
Saccharomyces cerevisiaeTSC11/AVO3 participates in regulating cell integrity and functionally interacts with components of the Tor2 complex.Curr Genet. 2005 May;47(5):273-88. doi: 10.1007/s00294-005-0570-8. Epub 2005 Apr 5. Curr Genet. 2005. PMID: 15809876
-
Molecular organization of target of rapamycin complex 2.J Biol Chem. 2005 Sep 2;280(35):30697-704. doi: 10.1074/jbc.M505553200. Epub 2005 Jul 7. J Biol Chem. 2005. PMID: 16002396
-
Regulation of TORC2 function and localization by Rab5 GTPases in Saccharomyces cerevisiae.Cell Cycle. 2019 May;18(10):1084-1094. doi: 10.1080/15384101.2019.1616999. Epub 2019 May 15. Cell Cycle. 2019. PMID: 31068077 Free PMC article. Review.
-
Insight into Tor2, a budding yeast microdomain protein.Eur J Cell Biol. 2014 Mar;93(3):87-97. doi: 10.1016/j.ejcb.2014.01.004. Epub 2014 Feb 14. Eur J Cell Biol. 2014. PMID: 24629393 Review.
Cited by
-
Target of rapamycin complex 2 signals to downstream effector yeast protein kinase 2 (Ypk2) through adheres-voraciously-to-target-of-rapamycin-2 protein 1 (Avo1) in Saccharomyces cerevisiae.J Biol Chem. 2012 Feb 24;287(9):6089-99. doi: 10.1074/jbc.M111.303701. Epub 2011 Dec 28. J Biol Chem. 2012. PMID: 22207764 Free PMC article.
-
Bioinformatic Analysis of Two TOR (Target of Rapamycin)-Like Proteins Encoded by Entamoeba histolytica Revealed Structural Similarities with Functional Homologs.Genes (Basel). 2021 Jul 28;12(8):1139. doi: 10.3390/genes12081139. Genes (Basel). 2021. PMID: 34440318 Free PMC article.
-
Comparative Proteomics Analysis Reveals Unique Early Signaling Response of Saccharomyces cerevisiae to Oxidants with Different Mechanism of Action.Int J Mol Sci. 2020 Dec 26;22(1):167. doi: 10.3390/ijms22010167. Int J Mol Sci. 2020. PMID: 33375274 Free PMC article.
-
TOR complex 2 is a master regulator of plasma membrane homeostasis.Biochem J. 2022 Sep 30;479(18):1917-1940. doi: 10.1042/BCJ20220388. Biochem J. 2022. PMID: 36149412 Free PMC article.
-
Life in the midst of scarcity: adaptations to nutrient availability in Saccharomyces cerevisiae.Curr Genet. 2010 Feb;56(1):1-32. doi: 10.1007/s00294-009-0287-1. Curr Genet. 2010. PMID: 20054690 Review.
References
-
- Alberts, A. S., N. Bouquin, L. H. Johnston, and R. Treisman. 1998. Analysis of RhoA-binding proteins reveals an interaction domain conserved in heterotrimeric G protein beta subunits and the yeast response regulator protein Skn7. J. Biol. Chem. 2738616-8622. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

