Expression of myogenic regulatory factors in the muscle-derived electric organ of Sternopygus macrurus
- PMID: 18552307
- PMCID: PMC2650510
- DOI: 10.1242/jeb.016592
Expression of myogenic regulatory factors in the muscle-derived electric organ of Sternopygus macrurus
Abstract
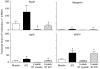
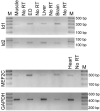
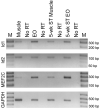
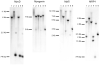
In most groups of electric fish, the current-producing cells of electric organs (EOs) derive from striated muscle fibers but retain some phenotypic characteristics of their precursor muscle cells. Given the role of the MyoD family of myogenic regulatory factors (MRFs) in the transcriptional activation of the muscle program in vertebrates, we examined their expression in the electrocytes of the gymnotiform Sternopygus macrurus. We estimated the number of MRF genes in the S. macrurus genome and our Southern blot analyses revealed a single MyoD, myogenin, myf5 and MRF4 gene. Quantitative RT-PCR showed that muscle and EO transcribe all MRF genes. With the exception of MyoD, the endogenous levels of myogenin, myf5 and MRF4 transcripts in electrocytes were greater than those detected in muscle fibers. These data indicate that MRF expression levels are not sufficient to predict the level to which the muscle program is manifested. Qualitative expression analysis of MRF co-regulators MEF2C, Id1 and Id2 also revealed these genes not to be unique to either muscle or EO, and detected similar expression patterns in the two tissues. Therefore, the partial muscle program of the EO is not associated with a partial expression of MRFs or with apparent distinct levels of some MRF co-factors. In addition, electrical inactivation by spinal cord transection (ST) resulted in the up-regulation of some muscle proteins in electrocytes without an accompanying increase in MRF transcript levels or notable changes in the co-factors MEF2C, Id1 and Id2. These findings suggest that the neural regulation of the skeletal muscle program via MRFs in S. macrurus might differ from that of their mammalian counterparts. Together, these data further our understanding of the molecular processes involved in the plasticity of the vertebrate skeletal muscle program that brings about the muscle-like phenotype of the non-contractile electrogenic cells in S. macrurus.
Figures



References
-
- Benezra R, Davis RL, Lockshon D, Turner DL, Weintraub H. The protein Id: a negative regulator of helix-loop-helix DNA binding proteins. Cell. 1990;61:49–59. - PubMed
-
- Braun T, Winter B, Bober E, Arnold HH. Transcriptional activation domain of the muscle-specific gene regulatory protein myf5. Nature. 1990;346:663–665. - PubMed
-
- Buckingham ME. Muscle: the regulation of myogenesis. Curr. Opin. Genet. Dev. 1994;4:745–751. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

