Tracking the complex flow of chromosome rearrangements from the Hominoidea Ancestor to extant Hylobates and Nomascus Gibbons by high-resolution synteny mapping
- PMID: 18552313
- PMCID: PMC2527702
- DOI: 10.1101/gr.078295.108
Tracking the complex flow of chromosome rearrangements from the Hominoidea Ancestor to extant Hylobates and Nomascus Gibbons by high-resolution synteny mapping
Abstract
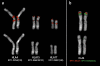
In this study we characterized the extension, reciprocal arrangement, and orientation of syntenic chromosomal segments in the lar gibbon (Hylobates lar, HLA) by hybridization of a panel of approximately 1000 human BAC clones. Each lar gibbon rearrangement was defined by a splitting BAC clone or by two overlapping clones flanking the breakpoint. A reconstruction of the synteny arrangement of the last common ancestor of all living lesser apes was made by combining these data with previous results in Nomascus leucogenys, Hoolock hoolock, and Symphalangus syndactylus. The definition of the ancestral synteny organization facilitated tracking the cascade of chromosomal changes from the Hominoidea ancestor to the present day karyotype of Hylobates and Nomascus. Each chromosomal rearrangement could be placed within an approximate phylogenetic and temporal framework. We identified 12 lar-specific rearrangements and five previously undescribed rearrangements that occurred in the Hylobatidae ancestor. The majority of the chromosomal differences between lar gibbons and humans are due to rearrangements that occurred in the Hylobatidae ancestor (38 events), consistent with the hypothesis that the genus Hylobates is the most recently evolved lesser ape genus. The rates of rearrangements in gibbons are 10 to 20 times higher than the mammalian default rate. Segmental duplication may be a driving force in gibbon chromosome evolution, because a consistent number of rearrangements involves pericentromeric regions (10 events) and centromere inactivation (seven events). Both phenomena can be reasonably supposed to have strongly contributed to the euchromatic dispersal of segmental duplications typical of pericentromeric regions. This hypothesis can be more fully tested when the sequence of this gibbon species becomes available. The detailed synteny map provided here will, in turn, substantially facilitate sequence assembly efforts.
Figures


Similar articles
-
A high-resolution map of synteny disruptions in gibbon and human genomes.PLoS Genet. 2006 Dec 29;2(12):e223. doi: 10.1371/journal.pgen.0020223. Epub 2006 Nov 13. PLoS Genet. 2006. PMID: 17196042 Free PMC article.
-
Chromosomal phylogeny and evolution of gibbons (Hylobatidae).Hum Genet. 2003 Nov;113(6):493-501. doi: 10.1007/s00439-003-0997-2. Epub 2003 Sep 3. Hum Genet. 2003. PMID: 14569461
-
Molecular refinement of gibbon genome rearrangements.Genome Res. 2007 Feb;17(2):249-57. doi: 10.1101/gr.6052507. Epub 2006 Dec 21. Genome Res. 2007. PMID: 17185643 Free PMC article.
-
Studies on karyotype evolution in higher primates in relation to human chromosome 14 and 9 by comparative mapping of immunoglobulin C epsilon genes with fluorescence in situ hybridization.Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 1999;(117):77-90. Kokuritsu Iyakuhin Shokuhin Eisei Kenkyusho Hokoku. 1999. PMID: 10859938 Review.
-
Fluorescence in situ hybridization to chromosomes as a tool to understand human and primate genome evolution.Cytogenet Genome Res. 2005;108(1-3):139-60. doi: 10.1159/000080811. Cytogenet Genome Res. 2005. PMID: 15545725 Review.
Cited by
-
Comparative genomics reveals birth and death of fragile regions in mammalian evolution.Genome Biol. 2010;11(11):R117. doi: 10.1186/gb-2010-11-11-r117. Epub 2010 Nov 30. Genome Biol. 2010. PMID: 21118492 Free PMC article.
-
Species identification of crested gibbons (Nomascus) in captivity in China using karyotyping- and PCR-based approaches.Zool Res. 2018 Sep 18;39(5):356-363. doi: 10.24272/j.issn.2095-8137.2018.036. Epub 2018 Apr 3. Zool Res. 2018. PMID: 29616678 Free PMC article.
-
Centromeric transposable elements and epigenetic status drive karyotypic variation in the eastern hoolock gibbon.Cell Genom. 2025 Apr 9;5(4):100808. doi: 10.1016/j.xgen.2025.100808. Epub 2025 Mar 14. Cell Genom. 2025. PMID: 40088887 Free PMC article.
-
The genome diversity and karyotype evolution of mammals.Mol Cytogenet. 2011 Oct 12;4:22. doi: 10.1186/1755-8166-4-22. Mol Cytogenet. 2011. PMID: 21992653 Free PMC article.
-
Patterns of genetic variation within and between Gibbon species.Mol Biol Evol. 2011 Aug;28(8):2211-8. doi: 10.1093/molbev/msr033. Epub 2011 Mar 2. Mol Biol Evol. 2011. PMID: 21368318 Free PMC article.
References
-
- Armengol L., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Pujana M.A., Cheung J., Scherer S.W., Estivill X., Cheung J., Scherer S.W., Estivill X., Scherer S.W., Estivill X., Estivill X. Enrichment of segmental duplications in regions of breaks of synteny between the human and mouse genomes suggest their involvement in evolutionary rearrangements. Hum. Mol. Genet. 2003;12:2201–2208. - PubMed
-
- Arnold N., Stanyon R., Jauch A., O'Brien P., Wienberg J., Stanyon R., Jauch A., O'Brien P., Wienberg J., Jauch A., O'Brien P., Wienberg J., O'Brien P., Wienberg J., Wienberg J. Identification of complex chromosome rearrangements in the gibbon by fluorescence in situ hybridization (FISH) of a human chromosome 2q specific microlibrary, yeast artificial chromosomes, and reciprocal chromosome painting. Cytogenet. Cell Genet. 1996;74:80–85. - PubMed
-
- Baena A., Mootnick A.R., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Mootnick A.R., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Falvo J.V., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Tsytskova A.V., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Ligeiro F., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Diop O.M., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Brieva C., Gagneux P., O'Brien S.J., Ryder O.A., Gagneux P., O'Brien S.J., Ryder O.A., O'Brien S.J., Ryder O.A., Ryder O.A., et al. Primate TNF promoters reveal markers of phylogeny and evolution of innate immunity. PLoS ONE. 2007;2:e621. doi: 10.1371/journal.pone.0000621. - DOI - PMC - PubMed
-
- Bailey W.J., Fitch D.H.A., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Fitch D.H.A., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Tagle D.A., Czelusniak J., Slightom J.L., Goodman M., Czelusniak J., Slightom J.L., Goodman M., Slightom J.L., Goodman M., Goodman M. Molecular evolution of the psi eta-globin gene locus: Gibbon phylogeny and the hominoid slowdown. Mol. Biol. Evol. 1991;8:155–184. - PubMed
-
- Bailey J.A., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Baertsch R., Kent W.J., Haussler D., Eichler E.E., Kent W.J., Haussler D., Eichler E.E., Haussler D., Eichler E.E., Eichler E.E. Hotspots of mammalian chromosomal evolution. Genome Biol. 2004a;5:R23. http://genomebiology.com/2004/5/4/R23 - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
