Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications
- PMID: 18552770
- PMCID: PMC2475317
- DOI: 10.1038/embor.2008.104
Bringing order to translation: the contributions of transfer RNA anticodon-domain modifications
Abstract
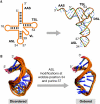
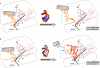
The biosynthesis of RNA includes its post-transcriptional modifications, and the crucial functions of these modifications have supported their conservation within all three kingdoms. For example, the modifications located within or adjacent to the anticodon of the transfer RNA (tRNA) enhance the accuracy of codon binding, maintain the translational reading frame and enable translocation of the tRNA from the A-site to the P-site of the ribosome. Although composed of different chemistries, the more than 70 known modifications of tRNA have in common their ability to reduce conformational dynamics, and to bring order to the internal loops and hairpin structures of RNA. The modified nucleosides of the anticodon domain of tRNA restrict its dynamics and shape its architecture; therefore, the need of the ribosome to constrain or remodel each tRNA to fit the decoding site is diminished. This concept reduces an entropic penalty for translation and provides a physicochemical basis for the conservation of RNA modifications in general.
Figures


References
-
- Agris PF (1991) Wobble position modified nucleosides evolved to select transfer RNA codon recognition: a modified-wobble hypothesis. Biochimie 73: 1345–1349 - PubMed
-
- Agris PF (1996) The importance of being modified: roles of modified nucleosides and Mg2+ in RNA structure and function. Progr Nucl Acid Res Mol Biol 53: 79–129 - PubMed
-
- Agris PF, Vendeix F, Graham WD (2007) tRNA's Wobble decoding of the genome: 40 years of modification. J Mol Biol 366: 1–13 - PubMed
-
- Ambrogelly A, Palioura S, Söll D (2007) Natural expansion of the genetic code. Nat Chem Biol 3: 29–35 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

