Genomic complexity of the Y-STR DYS19: inversions, deletions and founder lineages carrying duplications
- PMID: 18553096
- PMCID: PMC2680205
- DOI: 10.1007/s00414-008-0253-3
Genomic complexity of the Y-STR DYS19: inversions, deletions and founder lineages carrying duplications
Abstract
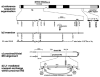
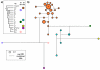
The Y-STR DYS19 is firmly established in the repertoire of Y-chromosomal markers used in forensic analysis yet is poorly understood at the molecular level, lying in a complex genomic environment and exhibiting null alleles, as well as duplications and occasional triplications in population samples. Here, we analyse three null alleles and 51 duplications and show that DYS19 can also be involved in inversion events, so that even its location within the short arm of the Y chromosome is uncertain. Deletion mapping in the three chromosomes carrying null alleles shows that their deletions are less than approximately 300 kb in size. Haplotypic analysis with binary markers shows that they belong to three different haplogroups and so represent independent events. In contrast, a collection of 51 DYS19 duplication chromosomes belong to only four haplogroups: two are singletons and may represent somatic mutation in lymphoblastoid cell lines, but two, in haplogroups G and C3c, represent founder lineages that have spread widely in Central Europe/West Asia and East Asia, respectively. Consideration of candidate mechanisms underlying both deletions and duplications provides no evidence for the involvement of non-allelic homologous recombination, and they are likely to represent sporadic events with low mutation rates. Understanding the basis and population distribution of these DYS19 alleles will aid in the utilisation and interpretation of profiles that contain them.
Figures
References
-
- Jobling MA, Pandya A, Tyler-Smith C. The Y chromosome in forensic analysis and paternity testing. Int J Legal Med. 1997;110:118–124. - PubMed
-
- Willuweit S, Roewer L, on behalf of the International Forensic Y Chromosome User Group Y chromosome haplotype reference database (YHRD): update. Forensic Sci Int Genet. 2007;1:83–87. - PubMed
-
- Roewer L, Arnemann J, Spurr NK, Grzeschik KH, Epplen JT. Simple repeat sequences on the human Y chromosome are equally polymorphic as their autosomal counterparts. Hum Genet. 1992;89:389–394. - PubMed
-
- Roewer L, Epplen JT. Rapid and sensitive typing of forensic stains by PCR amplification of polymorphic simple repeat sequences in case work. Forensic Sci Int. 1992;53:163–171. - PubMed
-
- Santos FR, Epplen JT, Pena SDJ. Testing deficiency paternity cases with a Y-linked tetranucleotide repeat polymorphism. In: Pena SDJ, Chakraborty R, Epplen JT, Jeffreys AJ, editors. DNA fingerprinting: state of the science. Birkhäuser Verlag; Basel: 1993. pp. 261–265. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

