Folding mechanisms of individual beta-hairpins in a Go model of Pin1 WW domain by all-atom molecular dynamics simulations
- PMID: 18554060
- PMCID: PMC2596927
- DOI: 10.1063/1.2936832
Folding mechanisms of individual beta-hairpins in a Go model of Pin1 WW domain by all-atom molecular dynamics simulations
Abstract
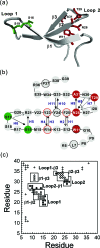
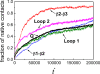
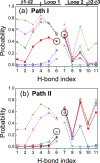
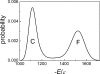
This paper examines the folding mechanism of an individual beta-hairpin in the presence of other hairpins by using an off-lattice model of a small triple-stranded antiparallel beta-sheet protein, Pin1 WW domain. The turn zipper model and the hydrophobic collapse model originally developed for a single beta-hairpin in literature is confirmed to be useful in describing beta-hairpins in model Pin1 WW domain. We find that the mechanism for folding a specific hairpin is independent of whether it folds first or second, but the formation process are significantly dependent on temperature. More specifically, beta1-beta2 hairpin folds via the turn zipper model at a low temperature and the hydrophobic collapse model at a high temperature, while the folding of beta2-beta3 hairpin follows the turn zipper model at both temperatures. The change in folding mechanisms is interpreted by the interplay between contact stability (enthalpy) and loop lengths (entropy), the effect of which is temperature dependent.
Figures



Similar articles
-
Temperature-dependent folding pathways of Pin1 WW domain: an all-atom molecular dynamics simulation of a Gō model.Biophys J. 2007 Sep 15;93(6):2152-61. doi: 10.1529/biophysj.106.102095. Epub 2007 May 18. Biophys J. 2007. PMID: 17513360 Free PMC article.
-
Engineering a beta-sheet protein toward the folding speed limit.J Phys Chem B. 2005 Aug 18;109(32):15182-6. doi: 10.1021/jp052373y. J Phys Chem B. 2005. PMID: 16852923
-
Analysis of PIN1 WW domain through a simple statistical mechanics model.Biophys Chem. 2005 Apr 1;115(2-3):153-8. doi: 10.1016/j.bpc.2004.12.020. Epub 2004 Dec 25. Biophys Chem. 2005. PMID: 15752598
-
Understanding the mechanism of beta-sheet folding from a chemical and biological perspective.Biopolymers. 2008;90(6):751-8. doi: 10.1002/bip.21101. Biopolymers. 2008. PMID: 18844292 Review.
-
Functions of WW domains in the nucleus.FEBS Lett. 2001 Feb 16;490(3):190-5. doi: 10.1016/s0014-5793(01)02122-6. FEBS Lett. 2001. PMID: 11223034 Review.
Cited by
-
Modeling the mechanism of CLN025 beta-hairpin formation.J Chem Phys. 2017 Sep 14;147(10):104107. doi: 10.1063/1.4993207. J Chem Phys. 2017. PMID: 28915754 Free PMC article.
-
Insights from coarse-grained Gō models for protein folding and dynamics.Int J Mol Sci. 2009 Mar;10(3):889-905. doi: 10.3390/ijms10030889. Epub 2009 Mar 2. Int J Mol Sci. 2009. PMID: 19399227 Free PMC article. Review.
-
Is there an en route folding intermediate for Cold shock proteins?Protein Sci. 2012 May;21(5):677-85. doi: 10.1002/pro.2053. Epub 2012 Mar 29. Protein Sci. 2012. PMID: 22467601 Free PMC article.
-
Biomolecular dynamics: order-disorder transitions and energy landscapes.Rep Prog Phys. 2012 Jul;75(7):076601. doi: 10.1088/0034-4885/75/7/076601. Epub 2012 Jun 28. Rep Prog Phys. 2012. PMID: 22790780 Free PMC article.
-
Investigating the Release of a Hydrophobic Peptide from Matrices of Biodegradable Polymers: An Integrated Method Approach.Polymer (Guildf). 2013 Jul 8;54(15):3806-3820. doi: 10.1016/j.polymer.2013.05.038. Polymer (Guildf). 2013. PMID: 24039300 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

