The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis
- PMID: 18554329
- PMCID: PMC2562793
- DOI: 10.1111/j.1365-2958.2008.06299.x
The specialized secretory apparatus ESX-1 is essential for DNA transfer in Mycobacterium smegmatis
Abstract
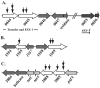
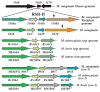
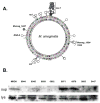
Conjugal DNA transfer in Mycobacterium smegmatis occurs by a mechanism distinct from plasmid-mediated DNA transfer. Previously, we had shown that the secretory apparatus, ESX-1, negatively regulated DNA transfer from the donor strain; ESX-1 donor mutants are hyper-conjugative. Here, we describe a genome-wide transposon mutagenesis screen to isolate recipient mutants. Surprisingly, we find that a majority of insertions map within the esx-1 locus, which encodes the secretory apparatus. Thus, in contrast to its role in donor function, ESX-1 is essential for recipient function; recipient ESX-1 mutants are hypo-conjugative. In addition to esx-1 genes, our screen identifies novel non-esx-1 loci in the M. smegmatis genome that are required for both DNA transfer and ESX-1 activity. DNA transfer therefore provides a simple molecular genetic assay to characterize ESX-1, which, in Mycobacterium tuberculosis, is necessary for full virulence. These findings reinforce the functional intertwining of DNA transfer and ESX-1 secretion, first described in the M. smegmatis donor. Moreover, our observation that ESX-1 has such diametrically opposed effects on transfer in the donor and recipient, forces us to consider how proteins secreted by the ESX-1 apparatus can function so as to modulate two seemingly disparate processes, M. smegmatis DNA transfer and M. tuberculosis virulence.
Figures

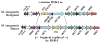
References
-
- Abdallah AM, Gey van Pittius NC, Champion PA, Cox J, Luirink J, Vandenbroucke-Grauls CM, Appelmelk BJ, Bitter W. Type VII secretion--mycobacteria show the way. Nat Rev Microbiol. 2007;5:883–891. - PubMed
-
- Bardarov S, Kriakov J, Carriere C, Yu S, Vaamonde C, McAdam RA, Bloom BR, Hatfull GF, Jacobs WRJ. Conditionally replicating mycobacteriophages: A system for transposon delivery to Mycobacterium tuberculosis. Proceedings of the National Academy of Sciences of the United States of America; 1997. pp. 10961–10966. - PMC - PubMed
-
- Bardarov S, Bardarov S, Jr, Pavelka MS, Jr, Sambandamurthy V, Larsen M, Tufariello J, Chan J, Hatfull G, Jacobs WR., Jr Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology. 2002;148:3007–3017. - PubMed
-
- Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science. 1999;284:1520–1523. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

