Identification of transcripts with enriched expression in the developing and adult pancreas
- PMID: 18554416
- PMCID: PMC2481431
- DOI: 10.1186/gb-2008-9-6-r99
Identification of transcripts with enriched expression in the developing and adult pancreas
Abstract
Background: Despite recent advances, the transcriptional hierarchy driving pancreas organogenesis remains largely unknown, in part due to the paucity of comprehensive analyses. To address this deficit we generated ten SAGE libraries from the developing murine pancreas spanning Theiler stages 17-26, making use of available Pdx1 enhanced green fluorescent protein (EGFP) and Neurog3 EGFP reporter strains, as well as tissue from adult islets and ducts.
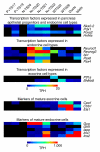
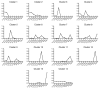
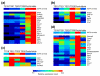
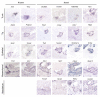
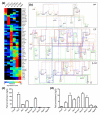
Results: We used a specificity metric to identify 2,536 tags with pancreas-enriched expression compared to 195 other mouse SAGE libraries. We subsequently grouped co-expressed transcripts with differential expression during pancreas development using K-means clustering. We validated the clusters first using quantitative real time PCR and then by analyzing the Theiler stage 22 pancreas in situ hybridization staining patterns of over 600 of the identified genes using the GenePaint database. These were then categorized into one of the five expression domains within the developing pancreas. Based on these results we identified a cascade of transcriptional regulators expressed in the endocrine pancreas lineage and, from this, we developed a predictive regulatory network describing beta-cell development.
Conclusion: Taken together, this work provides evidence that the SAGE libraries generated here are a valuable resource for continuing to elucidate the molecular mechanisms regulating pancreas development. Furthermore, our studies provide a comprehensive analysis of pancreas development, and insights into the regulatory networks driving this process are revealed.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

