An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors
- PMID: 18554609
- PMCID: PMC5367387
- DOI: 10.1016/j.jmb.2008.05.027
An indigenous posttranscriptional modification in the ribosomal peptidyl transferase center confers resistance to an array of protein synthesis inhibitors
Abstract
A number of nucleotide residues in ribosomal RNA (rRNA) undergo specific posttranscriptional modifications. The roles of most modifications are unclear, but their clustering in functionally important regions of rRNA suggests that they might either directly affect the activity of the ribosome or modulate its interactions with ligands. Of the 25 modified nucleotides in Escherichia coli 23S rRNA, 14 are located in the peptidyl transferase center, the main antibiotic target in the large ribosomal subunit. Since nucleotide modifications have been closely associated with both antibiotic sensitivity and antibiotic resistance, loss of some of these posttranscriptional modifications may affect the susceptibility of bacteria to antibiotics. We investigated the antibiotic sensitivity of E. coli cells in which the genes of 8 rRNA-modifying enzymes targeting the peptidyl transferase center were individually inactivated. The lack of pseudouridine at position 2504 of 23S rRNA was found to significantly increase the susceptibility of bacteria to peptidyl transferase inhibitors. Therefore, this indigenous posttranscriptional modification may have evolved as an intrinsic resistance mechanism protecting bacteria against natural antibiotics.
Figures
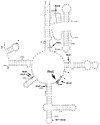

References
-
- Liang XH, Liu Q, Fournier MJ. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell. 2007;28:965–977. - PubMed
-
- Decatur WA, Liang XH, Piekna-Przybylska D, Fournier MJ. Identifying effects of snoRNA-guided modifications on the synthesis and function of the yeast ribosome. Methods Enzymol. 2007;425:283–316. - PubMed
-
- Xu Z, O'Farrell HC, Rife JP, Culver GM. A conserved rRNA methyltransferase regulates ribosome biogenesis. Nat. Struct. Mol. Biol. 2008;X XXX-XXX. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

