Natural selection on the Drosophila antimicrobial immune system
- PMID: 18555739
- PMCID: PMC2527063
- DOI: 10.1016/j.mib.2008.05.001
Natural selection on the Drosophila antimicrobial immune system
Abstract
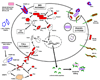
The evolutionary dynamics of immune defenses have long attracted interest because of the special role the immune system plays in mediating the antagonistic interaction between hosts and pathogens. The antimicrobial immune system of the fruit fly Drosophila melanogaster is genetically well characterized and serves as a valuable model for studying insect and human innate immune defenses. I review here evolutionary and comparative genomic analyses of insect antimicrobial immune genes, with an emphasis on Drosophila. Core signal transduction pathways in the immune system are orthologously conserved across long evolutionary distances, but genes in these pathways evolve rapidly and adaptively at the amino acid sequence level. By contrast, families of genes encoding antimicrobial peptides are remarkably dynamic in genomic duplication and deletion, yet individual genes show little indication of adaptive sequence evolution. Pattern recognition receptors that trigger humoral immunity are evolutionarily rather static, but receptors required for phagocytosis show considerable genomic rearrangement and adaptive sequence divergence. The distinct evolutionary patterns exhibited by these various classes of immune system genes can be logically connected to the functions of the proteins they encode.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

