Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase
- PMID: 18556022
- PMCID: PMC2556551
- DOI: 10.1016/j.jmb.2008.05.025
Structural and biochemical characterization of the therapeutic Anabaena variabilis phenylalanine ammonia lyase
Abstract
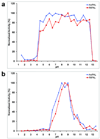
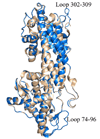
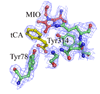
We have recently observed promising success in a mouse model for treating the metabolic disorder phenylketonuria with phenylalanine ammonia lyase (PAL) from Rhodosporidium toruloides and Anabaena variabilis. Both molecules, however, required further optimization in order to overcome problems with protease susceptibility, thermal stability, and aggregation. Previously, we optimized PAL from R. toruloides, and in this case we reduced aggregation of the A. variabilis PAL by mutating two surface cysteine residues (C503 and C565) to serines. Additionally, we report the structural and biochemical characterization of the A. variabilis PAL C503S/C565S double mutant and carefully compare this molecule with the R. toruloides engineered PAL molecule. Unlike previously published PAL structures, significant electron density is observed for the two active-site loops in the A. variabilis C503S/C565S double mutant, yielding a complete view of the active site. Docking studies and N-hydroxysuccinimide-biotin binding studies support a proposed mechanism in which the amino group of the phenylalanine substrate is attacked directly by the 4-methylidene-imidazole-5-one prosthetic group. We propose a helix-to-loop conformational switch in the helices flanking the inner active-site loop that regulates accessibility of the active site. Differences in loop stability among PAL homologs may explain the observed variation in enzyme efficiency, despite the highly conserved structure of the active site. A. variabilis C503S/C565S PAL is shown to be both more thermally stable and more resistant to proteolytic cleavage than R. toruloides PAL. Additional increases in thermal stability and protease resistance upon ligand binding may be due to enhanced interactions among the residues of the active site, possibly locking the active-site structure in place and stabilizing the tetramer. Examination of the A. variabilis C503S/C565S PAL structure, combined with analysis of its physical properties, provides a structural basis for further engineering of residues that could result in a better therapeutic molecule.
Figures


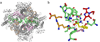

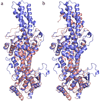
Similar articles
-
Directed evolution of Anabaena variabilis phenylalanine ammonia-lyase (PAL) identifies mutants with enhanced activities.Chem Commun (Camb). 2020 May 14;56(39):5255-5258. doi: 10.1039/d0cc00783h. Epub 2020 Apr 9. Chem Commun (Camb). 2020. PMID: 32270162 Free PMC article.
-
Modulating the pH Activity Profiles of Phenylalanine Ammonia Lyase from Anabaena variabilis by Modification of Center-Near Surface Residues.Appl Biochem Biotechnol. 2017 Nov;183(3):699-711. doi: 10.1007/s12010-017-2458-8. Epub 2017 Mar 25. Appl Biochem Biotechnol. 2017. PMID: 28343264
-
Preclinical evaluation of multiple species of PEGylated recombinant phenylalanine ammonia lyase for the treatment of phenylketonuria.Proc Natl Acad Sci U S A. 2008 Dec 30;105(52):20894-9. doi: 10.1073/pnas.0808421105. Epub 2008 Dec 18. Proc Natl Acad Sci U S A. 2008. PMID: 19095795 Free PMC article.
-
Biomedical applications of microbial phenylalanine ammonia lyase: Current status and future prospects.Biochimie. 2020 Oct;177:142-152. doi: 10.1016/j.biochi.2020.08.009. Epub 2020 Aug 21. Biochimie. 2020. PMID: 32828824 Review.
-
A modern view of phenylalanine ammonia lyase.Biochem Cell Biol. 2007 Jun;85(3):273-82. doi: 10.1139/o07-018. Biochem Cell Biol. 2007. PMID: 17612622 Review.
Cited by
-
X-Ray Crystallography in Structure-Function Characterization of Therapeutic Enzymes.Adv Exp Med Biol. 2019;1148:81-103. doi: 10.1007/978-981-13-7709-9_4. Adv Exp Med Biol. 2019. PMID: 31482495 Review.
-
Expression and properties of the highly alkalophilic phenylalanine ammonia-lyase of thermophilic Rubrobacter xylanophilus.PLoS One. 2014 Jan 27;9(1):e85943. doi: 10.1371/journal.pone.0085943. eCollection 2014. PLoS One. 2014. PMID: 24475062 Free PMC article.
-
A Methylidene Group in the Phosphonic Acid Analogue of Phenylalanine Reverses the Enantiopreference of Binding to Phenylalanine Ammonia-Lyases.Adv Synth Catal. 2017 Jun 19;359(12):2109-2120. doi: 10.1002/adsc.201700428. Epub 2017 May 19. Adv Synth Catal. 2017. PMID: 28919846 Free PMC article.
-
Directed evolution of Anabaena variabilis phenylalanine ammonia-lyase (PAL) identifies mutants with enhanced activities.Chem Commun (Camb). 2020 May 14;56(39):5255-5258. doi: 10.1039/d0cc00783h. Epub 2020 Apr 9. Chem Commun (Camb). 2020. PMID: 32270162 Free PMC article.
-
Clinical therapeutics for phenylketonuria.Drug Deliv Transl Res. 2012 Aug;2(4):223-37. doi: 10.1007/s13346-012-0067-1. Drug Deliv Transl Res. 2012. PMID: 25787029
References
-
- Scriver CR. In: The Metabolic and Molecular Bases of Inherited Disease. 8th edit. Scriver CR, editor. McGraw-Hill: Professional Publishing; 2000.
-
- Wang L, Gamez A, Sarkissian CN, Straub M, Patch MG, Han GW, Striepeke S, Fitzpatrick P, Scriver CR, Stevens RC. Structure-based chemical modification strategy for enzyme replacement treatment of phenylketonuria. Mol Genet Metab. 2005;86:134–140. - PubMed
-
- Hoskins JA, Jack G, Wade HE, Peiris RJ, Wright EC, Starr DJ, Stern J. Enzymatic control of phenylalanine intake in phenylketonuria. Lancet. 1980;1:392–394. - PubMed
-
- Ambrus CM, Anthone S, Horvath C, Kalghatgi K, Lele AS, Eapen G, Ambrus JL, Ryan AJ, Li P. Extracorporeal enzyme reactors for depletion of phenylalanine in phenylketonuria. Ann Intern Med. 1987;106:531–537. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

