Two variable active site residues modulate response regulator phosphoryl group stability
- PMID: 18557815
- PMCID: PMC2700761
- DOI: 10.1111/j.1365-2958.2008.06296.x
Two variable active site residues modulate response regulator phosphoryl group stability
Abstract
Many signal transduction networks control their output by switching regulatory elements on or off. To synchronize biological response with environmental stimulus, switching kinetics must be faster than changes in input. Two-component regulatory systems (used for signal transduction by bacteria, archaea and eukaryotes) switch via phosphorylation or dephosphorylation of the receiver domain in response regulator proteins. Although receiver domains share conserved active site residues and similar three-dimensional structures, rates of self-catalysed dephosphorylation span a >or= 40,000-fold range in response regulators that control diverse biological processes. For example, autodephosphorylation of the chemotaxis response regulator CheY is 640-fold faster than Spo0F, which controls sporulation. Here we demonstrate that substitutions at two variable active site positions decreased CheY autodephosphorylation up to 40-fold and increased the Spo0F rate up to 110-fold. Particular amino acids had qualitatively similar effects in different response regulators. However, mutant proteins matched to other response regulators at the two key variable positions did not always exhibit similar autodephosphorylation kinetics. Therefore, unknown factors also influence absolute rates. Understanding the effects that particular active site amino acid compositions have on autodephosphorylation rate may allow manipulation of phosphoryl group stability for useful purposes, as well as prediction of signal transduction kinetics from amino acid sequence.
Figures
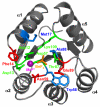
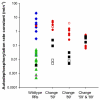


References
-
- Anderson JC, Clarke EJ, Arkin AP, Voigt CA. Environmentally controlled invasion of cancer cells by engineered bacteria. J Mol Biol. 2006;355:619–627. - PubMed
-
- Ashby MK. Survey of the number of two-component response regulator genes in the complete and annotated genome sequences of prokaryotes. FEMS Microbiol Lett. 2004;231:277–281. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

