Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant
- PMID: 18558690
- PMCID: PMC3982601
- DOI: 10.1021/ja711248y
Probing the substrate specificity of Golgi alpha-mannosidase II by use of synthetic oligosaccharides and a catalytic nucleophile mutant
Abstract
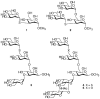
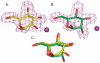
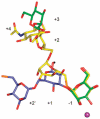
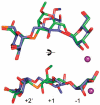
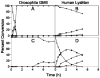
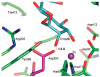
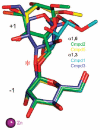
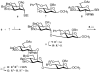
Inhibition of Golgi alpha-mannosidase II (GMII), which acts late in the N-glycan processing pathway, provides a route to blocking cancer-induced changes in cell surface oligosaccharide structures. To probe the substrate requirements of GMII, oligosaccharides were synthesized that contained an alpha(1,3)- or alpha(1,6)-linked 1-thiomannoside. Surprisingly, these oligosaccharides were not observed in X-ray crystal structures of native Drosophila GMII (dGMII). However, a mutant enzyme in which the catalytic nucleophilic aspartate was changed to alanine (D204A) allowed visualization of soaked oligosaccharides and led to the identification of the binding site for the alpha(1,3)-linked mannoside of the natural substrate. These studies also indicate that the conformational change of the bound mannoside to a high-energy B 2,5 conformation is facilitated by steric hindrance from, and the formation of strong hydrogen bonds to, Asp204. The observation that 1-thio-linked mannosides are not well tolerated by the catalytic site of dGMII led to the synthesis of a pentasaccharide containing the alpha(1,6)-linked Man of the natural substrate and the beta(1,2)-linked GlcNAc moiety proposed to be accommodated by the extended binding site of the enzyme. A cocrystal structure of this compound with the D204A enzyme revealed the molecular interactions with the beta(1,2)-linked GlcNAc. The structure is consistent with the approximately 80-fold preference of dGMII for the cleavage of substrates containing a nonreducing beta(1,2)-linked GlcNAc. By contrast, the lysosomal mannosidase lacks an equivalent GlcNAc binding site and kinetic analysis indicates oligomannoside substrates without non-reducing-terminal GlcNAc modifications are preferred, suggesting that selective inhibitors for GMII could exploit the additional binding specificity of the GlcNAc binding site.
Figures



Similar articles
-
Golgi alpha-mannosidase II cleaves two sugars sequentially in the same catalytic site.Proc Natl Acad Sci U S A. 2008 Jul 15;105(28):9570-5. doi: 10.1073/pnas.0802206105. Epub 2008 Jul 3. Proc Natl Acad Sci U S A. 2008. PMID: 18599462 Free PMC article.
-
Characterization of a cDNA encoding a novel human Golgi alpha 1, 2-mannosidase (IC) involved in N-glycan biosynthesis.J Biol Chem. 2000 Oct 13;275(41):31655-60. doi: 10.1074/jbc.M004935200. J Biol Chem. 2000. PMID: 10915796
-
Effect of substrate structure on the activity of Man9-mannosidase from pig liver involved in N-linked oligosaccharide processing.Eur J Biochem. 1992 Sep 1;208(2):451-7. doi: 10.1111/j.1432-1033.1992.tb17207.x. Eur J Biochem. 1992. PMID: 1521536
-
Golgi alpha-mannosidase II deficiency in vertebrate systems: implications for asparagine-linked oligosaccharide processing in mammals.Biochim Biophys Acta. 2002 Dec 19;1573(3):225-35. doi: 10.1016/s0304-4165(02)00388-4. Biochim Biophys Acta. 2002. PMID: 12417404 Review.
-
Structure, mechanism and inhibition of Golgi α-mannosidase II.Curr Opin Struct Biol. 2012 Oct;22(5):558-62. doi: 10.1016/j.sbi.2012.06.005. Epub 2012 Jul 20. Curr Opin Struct Biol. 2012. PMID: 22819743 Review.
Cited by
-
Characterisation of class I and II α-mannosidases from Drosophila melanogaster.Glycoconj J. 2013 Dec;30(9):899-909. doi: 10.1007/s10719-013-9495-5. Epub 2013 Aug 25. Glycoconj J. 2013. PMID: 23979800
-
N-Benzyl Substitution of Polyhydroxypyrrolidines: The Way to Selective Inhibitors of Golgi α-Mannosidase II.ChemMedChem. 2018 Feb 20;13(4):373-383. doi: 10.1002/cmdc.201700607. Epub 2018 Feb 6. ChemMedChem. 2018. PMID: 29323461 Free PMC article.
-
Structure and kinetic investigation of Streptococcus pyogenes family GH38 alpha-mannosidase.PLoS One. 2010 Feb 3;5(2):e9006. doi: 10.1371/journal.pone.0009006. PLoS One. 2010. PMID: 20140249 Free PMC article.
-
Glycan fragment database: a database of PDB-based glycan 3D structures.Nucleic Acids Res. 2013 Jan;41(Database issue):D470-4. doi: 10.1093/nar/gks987. Epub 2012 Oct 26. Nucleic Acids Res. 2013. PMID: 23104379 Free PMC article.
-
Engineering β1,4-galactosyltransferase I to reduce secretion and enhance N-glycan elongation in insect cells.J Biotechnol. 2015 Jan 10;193:52-65. doi: 10.1016/j.jbiotec.2014.11.013. Epub 2014 Nov 25. J Biotechnol. 2015. PMID: 25462875 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

