Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system
- PMID: 18558867
- PMCID: PMC2728073
- DOI: 10.1146/annurev.neuro.31.060407.125631
Signaling mechanisms linking neuronal activity to gene expression and plasticity of the nervous system
Abstract
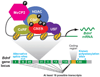
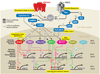
Sensory experience and the resulting synaptic activity within the brain are critical for the proper development of neural circuits. Experience-driven synaptic activity causes membrane depolarization and calcium influx into select neurons within a neural circuit, which in turn trigger a wide variety of cellular changes that alter the synaptic connectivity within the neural circuit. One way in which calcium influx leads to the remodeling of synapses made by neurons is through the activation of new gene transcription. Recent studies have identified many of the signaling pathways that link neuronal activity to transcription, revealing both the transcription factors that mediate this process and the neuronal activity-regulated genes. These studies indicate that neuronal activity regulates a complex program of gene expression involved in many aspects of neuronal development, including dendritic branching, synapse maturation, and synapse elimination. Genetic mutations in several key regulators of activity-dependent transcription give rise to neurological disorders in humans, suggesting that future studies of this gene expression program will likely provide insight into the mechanisms by which the disruption of proper synapse development can give rise to a variety of neurological disorders.
Figures

Similar articles
-
Communication between the synapse and the nucleus in neuronal development, plasticity, and disease.Annu Rev Cell Dev Biol. 2008;24:183-209. doi: 10.1146/annurev.cellbio.24.110707.175235. Annu Rev Cell Dev Biol. 2008. PMID: 18616423 Free PMC article. Review.
-
N-methyl-D-aspartate-stimulated ERK1/2 signaling and the transcriptional up-regulation of plasticity-related genes are developmentally regulated following in vitro neuronal maturation.J Neurosci Res. 2009 Sep;87(12):2632-44. doi: 10.1002/jnr.22103. J Neurosci Res. 2009. PMID: 19396876 Free PMC article.
-
Regulation of cpg15 by signaling pathways that mediate synaptic plasticity.Mol Cell Neurosci. 2003 Nov;24(3):538-54. doi: 10.1016/s1044-7431(03)00230-6. Mol Cell Neurosci. 2003. PMID: 14664806 Free PMC article.
-
Normal dendrite growth in Drosophila motor neurons requires the AP-1 transcription factor.Dev Neurobiol. 2008 Sep 1;68(10):1225-42. doi: 10.1002/dneu.20655. Dev Neurobiol. 2008. PMID: 18548486 Free PMC article.
-
Neuronal activity-regulated gene transcription in synapse development and cognitive function.Cold Spring Harb Perspect Biol. 2011 Jun 1;3(6):a005744. doi: 10.1101/cshperspect.a005744. Cold Spring Harb Perspect Biol. 2011. PMID: 21555405 Free PMC article. Review.
Cited by
-
Regulation of Cardiac Calcium Channels in the Fight-or-Flight Response.Curr Mol Pharmacol. 2015;8(1):12-21. doi: 10.2174/1874467208666150507103417. Curr Mol Pharmacol. 2015. PMID: 25966697 Free PMC article. Review.
-
Genome-Wide Analysis of Polyadenylation Events in Schmidtea mediterranea.G3 (Bethesda). 2016 Oct 13;6(10):3035-3048. doi: 10.1534/g3.116.031120. G3 (Bethesda). 2016. PMID: 27489207 Free PMC article.
-
Activity-Dependent Differential Regulation of Auts2 Isoforms In Vitro and In Vivo.Mol Neurobiol. 2023 Jun;60(6):2973-2985. doi: 10.1007/s12035-023-03241-x. Epub 2023 Feb 9. Mol Neurobiol. 2023. PMID: 36754912
-
Immediate-Early Promoter-Driven Transgenic Reporter System for Neuroethological Research in a Hemimetabolous Insect.eNeuro. 2018 Sep 4;5(4):ENEURO.0061-18.2018. doi: 10.1523/ENEURO.0061-18.2018. eCollection 2018 Jul-Aug. eNeuro. 2018. PMID: 30225346 Free PMC article.
-
In silico approach to calculate the transcript capacity.Genomics Inform. 2019 Sep;17(3):e31. doi: 10.5808/GI.2019.17.3.e31. Epub 2019 Sep 26. Genomics Inform. 2019. PMID: 31610627 Free PMC article.
References
-
- Aizawa H, Hu SC, Bobb K, Balakrishnan K, Ince G, et al. Dendrite development regulated by CREST, a calcium-regulated transcriptional activator. Science. 2004;303:197–202. - PubMed
-
- Bailey CH, Chen M. Morphological basis of long-term habituation and sensitization in Aplysia. Science. 1983;220:91–93. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

