Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium
- PMID: 18559084
- PMCID: PMC2443395
- DOI: 10.1186/1471-2164-9-291
Genome-wide transcriptional changes induced by phagocytosis or growth on bacteria in Dictyostelium
Abstract
Background: Phagocytosis plays a major role in the defense of higher organisms against microbial infection and provides also the basis for antigen processing in the immune response. Cells of the model organism Dictyostelium are professional phagocytes that exploit phagocytosis of bacteria as the preferred way to ingest food, besides killing pathogens. We have investigated Dictyostelium differential gene expression during phagocytosis of non-pathogenic bacteria, using DNA microarrays, in order to identify molecular functions and novel genes involved in phagocytosis.
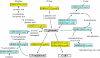
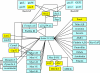
Results: The gene expression profiles of cells incubated for a brief time with bacteria were compared with cells either incubated in axenic medium or growing on bacteria. Transcriptional changes during exponential growth in axenic medium or on bacteria were also compared. We recognized 443 and 59 genes that are differentially regulated by phagocytosis or by the different growth conditions (growth on bacteria vs. axenic medium), respectively, and 102 genes regulated by both processes. Roughly one third of the genes are up-regulated compared to macropinocytosis and axenic growth. Functional annotation of differentially regulated genes with different tools revealed that phagocytosis induces profound changes in carbohydrate, amino acid and lipid metabolism, and in cytoskeletal components. Genes regulating translation and mitochondrial biogenesis are mostly up-regulated. Genes involved in sterol biosynthesis are selectively up-regulated, suggesting a shift in membrane lipid composition linked to phagocytosis. Very few changes were detected in genes required for vesicle fission/fusion, indicating that the intracellular traffic machinery is mostly in common between phagocytosis and macropinocytosis. A few putative receptors, including GPCR family 3 proteins, scaffolding and adhesion proteins, components of signal transduction and transcription factors have been identified, which could be part of a signalling complex regulating phagocytosis and adaptational downstream responses.
Conclusion: The results highlight differences between phagocytosis and macropinocytosis, and provide the basis for targeted functional analysis of new candidate genes and for comparison studies with transcriptomes during infection with pathogenic bacteria.
Figures


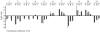



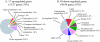


References
-
- Botelho RJ, Scott CC, Grinstein S. Phosphoinositide involvement in phagocytosis and phagosome maturation. Curr Top Microbiol Immunol. 2004;282:1–30. - PubMed
-
- Peracino B, Borleis J, Jin T, Westphal M, Schwartz JM, Wu L, Bracco E, Gerisch G, Devreotes P, Bozzaro S. G protein beta subunit-null mutants are impaired in phagocytosis and chemotaxis due to inappropriate regulation of the actin cytoskeleton. J Cell Biol. 1998;141:1529–1537. doi: 10.1083/jcb.141.7.1529. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

