Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha
- PMID: 18559093
- PMCID: PMC2440550
- DOI: 10.1186/1476-4598-7-58
Integrative genomics analysis of chromosome 5p gain in cervical cancer reveals target over-expressed genes, including Drosha
Abstract
Background: Copy number gains and amplifications are characteristic feature of cervical cancer (CC) genomes for which the underlying mechanisms are unclear. These changes may possess oncogenic properties by deregulating tumor-related genes. Gain of short arm of chromosome 5 (5p) is the most frequent karyotypic change in CC.
Methods: To examine the role of 5p gain, we performed a combination of single nucleotide polymorphism (SNP) array, fluorescence in situ hybridization (FISH), and gene expression analyses on invasive cancer and in various stages of CC progression.
Results: The SNP and FISH analyses revealed copy number increase (CNI) of 5p in 63% of invasive CC, which arises at later stages of precancerous lesions in CC development. We integrated chromosome 5 genomic copy number and gene expression data to identify key target over expressed genes as a consequence of 5p gain. One of the candidates identified was Drosha (RNASEN), a gene that is required in the first step of microRNA (miRNA) processing in the nucleus. Other 5p genes identified as targets of CNI play a role in DNA repair and cell cycle regulation (BASP1, TARS, PAIP1, BRD9, RAD1, SKP2, and POLS), signal transduction (OSMR), and mitochondrial oxidative phosphorylation (NNT, SDHA, and NDUFS6), suggesting that disruption of pathways involving these genes may contribute to CC progression.
Conclusion: Taken together, we demonstrate the power of integrating genomics data with expression data in deciphering tumor-related targets of CNI. Identification of 5p gene targets in CC denotes an important step towards biomarker development and forms a framework for testing as molecular therapeutic targets.
Figures
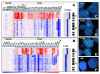
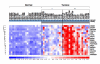

Similar articles
-
Identification of copy number gain and overexpressed genes on chromosome arm 20q by an integrative genomic approach in cervical cancer: potential role in progression.Genes Chromosomes Cancer. 2008 Sep;47(9):755-65. doi: 10.1002/gcc.20577. Genes Chromosomes Cancer. 2008. PMID: 18506748
-
Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article.
-
Identification of SEC62 as a potential marker for 3q amplification and cellular migration in dysplastic cervical lesions.BMC Cancer. 2016 Aug 23;16(1):676. doi: 10.1186/s12885-016-2739-6. BMC Cancer. 2016. PMID: 27553742 Free PMC article.
-
Integrative genomic approaches in cervical cancer: implications for molecular pathogenesis.Future Oncol. 2010 Oct;6(10):1643-52. doi: 10.2217/fon.10.114. Future Oncol. 2010. PMID: 21062161 Free PMC article. Review.
-
Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions.Nat Rev Cancer. 2014 Jun;14(6):395-405. doi: 10.1038/nrc3728. Nat Rev Cancer. 2014. PMID: 24854082 Review.
Cited by
-
LP99: Discovery and Synthesis of the First Selective BRD7/9 Bromodomain Inhibitor.Angew Chem Weinheim Bergstr Ger. 2015 May 18;127(21):6315-6319. doi: 10.1002/ange.201501394. Epub 2015 Apr 13. Angew Chem Weinheim Bergstr Ger. 2015. PMID: 27346896 Free PMC article.
-
Variability in the incidence of miRNAs and genes in fragile sites and the role of repeats and CpG islands in the distribution of genetic material.PLoS One. 2010 Jun 17;5(6):e11166. doi: 10.1371/journal.pone.0011166. PLoS One. 2010. PMID: 20567512 Free PMC article.
-
Murine Oncostatin M Has Opposing Effects on the Proliferation of OP9 Bone Marrow Stromal Cells and NIH/3T3 Fibroblasts Signaling through the OSMR.Int J Mol Sci. 2021 Oct 28;22(21):11649. doi: 10.3390/ijms222111649. Int J Mol Sci. 2021. PMID: 34769079 Free PMC article.
-
Genome-wide DNA methylation signatures to predict pathologic complete response from combined neoadjuvant chemotherapy with bevacizumab in breast cancer.PLoS One. 2020 Apr 16;15(4):e0230248. doi: 10.1371/journal.pone.0230248. eCollection 2020. PLoS One. 2020. PMID: 32298288 Free PMC article. Clinical Trial.
-
The genetic alteration spectrum of the SWI/SNF complex: The oncogenic roles of BRD9 and ACTL6A.PLoS One. 2019 Sep 10;14(9):e0222305. doi: 10.1371/journal.pone.0222305. eCollection 2019. PLoS One. 2019. PMID: 31504061 Free PMC article.
References
-
- Kirchhoff M, Rose H, Petersen BL, Maahr J, Gerdes T, Lundsteen C, Bryndorf T, Kryger-Baggesen N, Christensen L, Engelholm SA, Philip J. Comparative genomic hybridization reveals a recurrent pattern of chromosomal aberrations in severe dysplasia/carcinoma in situ of the cervix and in advanced-stage cervical carcinoma. Genes Chromosomes Cancer. 1999;24:144–150. doi: 10.1002/(SICI)1098-2264(199902)24:2<144::AID-GCC7>3.0.CO;2-9. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

