Relative effects of mutability and selection on single nucleotide polymorphisms in transcribed regions of the human genome
- PMID: 18559102
- PMCID: PMC2442617
- DOI: 10.1186/1471-2164-9-292
Relative effects of mutability and selection on single nucleotide polymorphisms in transcribed regions of the human genome
Abstract
Motivation: Single nucleotide polymorphisms (SNPs) are the most common type of genetic variation in humans. However, the factors that affect SNP density are poorly understood. The goal of this study was to estimate the relative effects of mutability and selection on SNP density in transcribed regions of human genes. It is important for prediction of the regions that harbor functional polymorphisms.
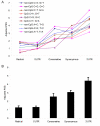
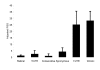
Results: We used frequency-validated SNPs resulting from single-nucleotide substitutions. SNPs were subdivided into five functional categories: (i) 5' untranslated region (UTR) SNPs, (ii) 3' UTR SNPs, (iii) synonymous SNPs, (iv) SNPs producing conservative missense mutations, and (v) SNPs producing radical missense mutations. Each of these categories was further subdivided into nine mutational categories on the basis of the single-nucleotide substitution type. Thus, 45 functional/mutational categories were analyzed. The relative mutation rate in each mutational category was estimated on the basis of published data. The proportion of segregating sites (PSSs) for each functional/mutational category was estimated by dividing the observed number of SNPs by the number of potential sites in the genome for a given functional/mutational category. By analyzing each functional group separately, we found significant positive correlations between PSSs and relative mutation rates (Spearman's correlation coefficient, at least r = 0.96, df = 9, P < 0.001). We adjusted the PSSs for the mutation rate and found that the functional category had a significant effect on SNP density (F = 5.9, df = 4, P = 0.001), suggesting that selection affects SNP density in transcribed regions of the genome. We used analyses of variance and covariance to estimate the relative effects of selection (functional category) and mutability (relative mutation rate) on the PSSs and found that approximately 87% of variation in PSS was due to variation in the mutation rate and approximately 13% was due to selection, suggesting that the probability that a site located in a transcribed region of a gene is polymorphic mostly depends on the mutability of the site.
Figures
Similar articles
-
Strand bias in complementary single-nucleotide polymorphisms of transcribed human sequences: evidence for functional effects of synonymous polymorphisms.BMC Genomics. 2006 Aug 17;7:213. doi: 10.1186/1471-2164-7-213. BMC Genomics. 2006. PMID: 16916449 Free PMC article.
-
Investigating single nucleotide polymorphism (SNP) density in the human genome and its implications for molecular evolution.Gene. 2003 Jul 17;312:207-13. doi: 10.1016/s0378-1119(03)00670-x. Gene. 2003. PMID: 12909357
-
Strength of the purifying selection against different categories of the point mutations in the coding regions of the human genome.Hum Mol Genet. 2006 Apr 1;15(7):1143-50. doi: 10.1093/hmg/ddl029. Epub 2006 Feb 24. Hum Mol Genet. 2006. PMID: 16500998
-
Prioritization of candidate SNPs in colon cancer using bioinformatics tools: an alternative approach for a cancer biologist.Interdiscip Sci. 2010 Dec;2(4):320-46. doi: 10.1007/s12539-010-0003-3. Epub 2010 Dec 12. Interdiscip Sci. 2010. PMID: 21153778 Review.
-
The ApoE gene of Alzheimer's disease (AD).Funct Integr Genomics. 2011 Dec;11(4):519-22. doi: 10.1007/s10142-011-0238-z. Epub 2011 Jul 19. Funct Integr Genomics. 2011. PMID: 21769591 Review.
Cited by
-
Mutability of druggable kinases and pro-inflammatory cytokines by their proximity to telomeres and A+T content.PLoS One. 2023 Apr 27;18(4):e0283470. doi: 10.1371/journal.pone.0283470. eCollection 2023. PLoS One. 2023. PMID: 37104389 Free PMC article.
-
Finding driver mutations in cancer: Elucidating the role of background mutational processes.PLoS Comput Biol. 2019 Apr 29;15(4):e1006981. doi: 10.1371/journal.pcbi.1006981. eCollection 2019 Apr. PLoS Comput Biol. 2019. PMID: 31034466 Free PMC article.
-
Unexpectedly low mutation rates in beta-myosin heavy chain and cardiac myosin binding protein genes in Italian patients with hypertrophic cardiomyopathy.J Cell Physiol. 2011 Nov;226(11):2894-900. doi: 10.1002/jcp.22636. J Cell Physiol. 2011. PMID: 21302287 Free PMC article.
-
Paternal age effect mutations and selfish spermatogonial selection: causes and consequences for human disease.Am J Hum Genet. 2012 Feb 10;90(2):175-200. doi: 10.1016/j.ajhg.2011.12.017. Am J Hum Genet. 2012. PMID: 22325359 Free PMC article. Review.
-
Evolutionary evidence of the effect of rare variants on disease etiology.Clin Genet. 2011 Mar;79(3):199-206. doi: 10.1111/j.1399-0004.2010.01535.x. Epub 2010 Sep 10. Clin Genet. 2011. PMID: 20831747 Free PMC article. Review.
References
-
- Collins FS, Guyer MS, Charkravarti A. Variations on a theme: cataloging human DNA sequence variation. Science. 1997;278:1580–1581. - PubMed
-
- Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nat Genet. 1999;22:231–238. - PubMed
-
- Long AD, Grote MN, Langley CH. Genetic analysis of complex diseases. Science. 1997;275:1328. author reply 1329-1330. - PubMed
-
- Miller MP, Kumar S. Understanding human disease mutations through the use of interspecific genetic variation. Hum Mol Genet. 2001;10:2319–2328. - PubMed
-
- Reich DE, Lander ES. On the allelic spectrum of human disease. Trends Genet. 2001;17:502–510. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous