Recombination in feline immunodeficiency virus from feral and companion domestic cats
- PMID: 18559113
- PMCID: PMC2453118
- DOI: 10.1186/1743-422X-5-76
Recombination in feline immunodeficiency virus from feral and companion domestic cats
Abstract
Background: Recombination is a relatively common phenomenon in retroviruses. We investigated recombination in Feline Immunodeficiency Virus from naturally-infected New Zealand domestic cats (Felis catus) by sequencing regions of the gag, pol and env genes.
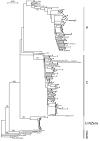
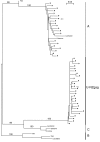
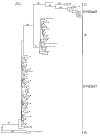
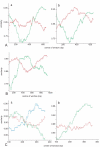
Results: The occurrence of intragenic recombination was highest in env, with evidence of recombination in 6.4% (n = 156) of all cats. A further recombinant was identified in each of the gag (n = 48) and pol (n = 91) genes. Comparisons of phylogenetic trees across genes identified cases of incongruence, indicating intergenic recombination. Three (7.7%, n = 39) of these incongruencies were found to be significantly different using the Shimodaira-Hasegawa test.Surprisingly, our phylogenies from the gag and pol genes showed that no New Zealand sequences group with reference subtype C sequences within intrasubtype pairwise distances. Indeed, we find one and two distinct unknown subtype groups in gag and pol, respectively. These observations cause us to speculate that these New Zealand FIV strains have undergone several recombination events between subtype A parent strains and undefined unknown subtype strains, similar to the evolutionary history hypothesised for HIV-1 "subtype E".Endpoint dilution sequencing was used to confirm the consensus sequences of the putative recombinants and unknown subtype groups, providing evidence for the authenticity of these sequences. Endpoint dilution sequencing also resulted in the identification of a dual infection event in the env gene. In addition, an intrahost recombination event between variants of the same subtype in the pol gene was established. This is the first known example of naturally-occurring recombination in a cat with infection of the parent strains.
Conclusion: Evidence of intragenic recombination in the gag, pol and env regions, and complex intergenic recombination, of FIV from naturally-infected domestic cats in New Zealand was found. Strains of unknown subtype were identified in all three gene regions. These results have implications for the use of the current FIV vaccine in New Zealand.
Figures

Similar articles
-
Molecular epidemiology of feline immunodeficiency virus in the domestic cat (Felis catus).Vet Immunol Immunopathol. 2010 Mar 15;134(1-2):68-74. doi: 10.1016/j.vetimm.2009.10.011. Epub 2009 Oct 14. Vet Immunol Immunopathol. 2010. PMID: 19896220 Free PMC article. Review.
-
Molecular subtyping of feline immunodeficiency virus from cats in Melbourne.Aust Vet J. 2008 Oct;86(10):385-9. doi: 10.1111/j.1751-0813.2008.00336.x. Aust Vet J. 2008. PMID: 18826508
-
Phylogeographic patterns of feline immunodeficiency virus genetic diversity in the domestic cat.Virology. 1998 Nov 25;251(2):234-43. doi: 10.1006/viro.1998.9402. Virology. 1998. PMID: 9837787
-
Phylogenetic analysis of feline immunodeficiency virus strains from naturally infected cats in Belgium and The Netherlands.Virus Res. 2015 Jan 22;196:30-6. doi: 10.1016/j.virusres.2014.10.023. Epub 2014 Nov 1. Virus Res. 2015. PMID: 25449575
-
Evolution of feline immunodeficiency virus in Felidae: implications for human health and wildlife ecology.Vet Immunol Immunopathol. 2008 May 15;123(1-2):32-44. doi: 10.1016/j.vetimm.2008.01.010. Epub 2008 Jan 19. Vet Immunol Immunopathol. 2008. PMID: 18359092 Free PMC article. Review.
Cited by
-
Molecular epidemiology of feline immunodeficiency virus in the domestic cat (Felis catus).Vet Immunol Immunopathol. 2010 Mar 15;134(1-2):68-74. doi: 10.1016/j.vetimm.2009.10.011. Epub 2009 Oct 14. Vet Immunol Immunopathol. 2010. PMID: 19896220 Free PMC article. Review.
-
Decreased Sensitivity of the Serological Detection of Feline Immunodeficiency Virus Infection Potentially Due to Imported Genetic Variants.Viruses. 2019 Jul 31;11(8):697. doi: 10.3390/v11080697. Viruses. 2019. PMID: 31370217 Free PMC article.
-
Prevalence of feline immunodeficiency virus and feline leukaemia virus in domestic cats in Hungary.JFMS Open Rep. 2019 Dec 10;5(2):2055116919892094. doi: 10.1177/2055116919892094. eCollection 2019 Jul-Dec. JFMS Open Rep. 2019. PMID: 31839979 Free PMC article.
-
Feline immunodeficiency virus (FIV) vaccine efficacy and FIV neutralizing antibodies.Vaccine. 2014 Feb 3;32(6):746-54. doi: 10.1016/j.vaccine.2013.05.024. Epub 2013 Jun 22. Vaccine. 2014. PMID: 23800540 Free PMC article.
-
Feline immunodeficiency virus in Northern Ceará, Brazil.JFMS Open Rep. 2019 Jul 8;5(2):2055116919859112. doi: 10.1177/2055116919859112. eCollection 2019 Jul-Dec. JFMS Open Rep. 2019. PMID: 31312511 Free PMC article.
References
-
- Troyer JL, Pecon-Slattery J, Roelke ME, Johnson W, VandeWoude S, Vazquez-Salat N, Brown M, Frank L, Woodroffe R, Winterbach C, Winterbach H, Hemson G, Bush M, Alexander KA, Revilla E, O'Brien SJ. Seroprevalence and genomic divergence of circulating strains of feline immunodeficiency virus among Felidae and Hyaenidae species. J Virol. 2005;79:8282–8294. doi: 10.1128/JVI.79.13.8282-8294.2005. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

