Primase-based whole genome amplification
- PMID: 18559358
- PMCID: PMC2490742
- DOI: 10.1093/nar/gkn377
Primase-based whole genome amplification
Abstract
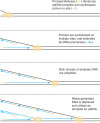
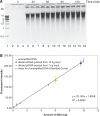
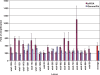
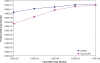
In vitro DNA amplification methods, such as polymerase chain reaction (PCR), rely on synthetic oligonucleotide primers for initiation of the reaction. In vivo, primers are synthesized on-template by DNA primase. The bacteriophage T7 gene 4 protein (gp4) has both primase and helicase activities. In this study, we report the development of a primase-based Whole Genome Amplification (pWGA) method, which utilizes gp4 primase to synthesize primers, eliminating the requirement of adding synthetic primers. Typical yield of pWGA from 1 ng to 10 ng of human genomic DNA input is in the microgram range, reaching over a thousand-fold amplification after 1 h of incubation at 37 degrees C. The amplification bias on human genomic DNA is 6.3-fold among 20 loci on different chromosomes. In addition to amplifying total genomic DNA, pWGA can also be used for detection and quantification of contaminant DNA in a sample when combined with a fluorescent reporter dye. When circular DNA is used as template in pWGA, 10(8)-fold of amplification is observed from as low as 100 copies of input. The high efficiency of pWGA in amplifying circular DNA makes it a potential tool in diagnosis and genotyping of circular human DNA viruses such as human papillomavirus (HPV).
Figures

References
-
- Telenius H, Carter NP, Bebb CE, Nordenskjold M, Ponder BA, Tunnacliffe A. Degenerate oligonucleotide-primed PCR: general amplification of target DNA by a single degenerate primer. Genomics. 1992;13:718–725. - PubMed
-
- Kittler R, Stoneking M, Kayser M. A whole genome amplification method to generate long fragments from low quantities of genomic DNA. Anal. Biochem. 2002;300:237–244. - PubMed
-
- Wells D, Delhanty JD. Comprehensive chromosomal analysis of human preimplantation embryos using whole genome amplification and single cell comparative genomic hybridization. Mol. Hum. Reprod. 2000;6:1055–1062. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

