Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis
- PMID: 18559475
- PMCID: PMC2428057
- DOI: 10.1101/gad.1671708
Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis
Abstract
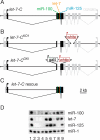
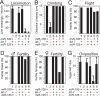
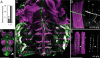
The Drosophila let-7-Complex (let-7-C) is a polycistronic locus encoding three ancient microRNAs: let-7, miR-100, and fly lin-4 (miR-125). We find that the let-7-C locus is principally expressed in the pupal and adult neuromusculature. let-7-C knockout flies appear normal externally but display defects in adult behaviors (e.g., flight, motility, and fertility) as well as clear juvenile features in their neuromusculature. We find that the function of let-7-C to ensure the appropriate remodeling of the abdominal neuromusculature during the larval-to-adult transition is carried out predominantly by let-7 alone. This heterochronic role of let-7 is likely just one of the ways in which let-7-C promotes adult behavior.
Figures


Comment in
-
A matter of timing: microRNA-controlled temporal identities in worms and flies.Genes Dev. 2008 Jun 15;22(12):1572-6. doi: 10.1101/gad.1690608. Genes Dev. 2008. PMID: 18559473 Free PMC article.
References
-
- Ambros V., Horvitz R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. - PubMed
-
- Bashirullah A., Pasquinelli A.E., Kiger A.A., Perrimon N., Ruvkun G., Thummel C.S. Coordinate regulation of small temporal RNAs at the onset of Drosophila metamorphosis. Dev. Biol. 2003;259:1–8. - PubMed
-
- Crossley A.C. The morphology and development of the Drosophila muscular system. In: Ashburner M., Wright T.R.F., editors. The genetics and biology of Drosophila. Academic Press; London: 1978. pp. 499–560.
-
- Currie D.A., Bate M. The development of adult abdominal muscles in Drosophila: Myoblasts express twist and are associated with nerves. Development. 1991;113:91–102. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
