Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading
- PMID: 18559476
- PMCID: PMC2428058
- DOI: 10.1101/gad.1667808
Tandem repeats upstream of the Arabidopsis endogene SDC recruit non-CG DNA methylation and initiate siRNA spreading
Abstract
Plants use siRNAs to target cytosine DNA methylation to both symmetrical CG and nonsymmetrical (CHG and CHH) sequence contexts. DNA methylation and siRNA clusters most frequently overlap with transposons in the Arabidopsis thaliana genome. However, a significant number of protein-coding genes also show promoter DNA methylation, and this can be used to silence their expression. Loss of the majority of non-CG DNA methylation in drm1 drm2 cmt3 triple mutants leads to developmental phenotypes. We identified the gene responsible for these phenotypes as SUPPRESSOR OF drm1 drm2 cmt3 (SDC), which encodes an F-box protein and possesses seven promoter tandem repeats. The SDC repeats show a unique silencing requirement for non-CG DNA methylation directed redundantly by histone methylation and siRNAs, and display spreading of siRNAs and methylation beyond the repeated region. In addition to revealing the complexity of DNA methylation control in A. thaliana, SDC has important implications for how plant genomes utilize gene silencing to repress endogenous genes.
Figures

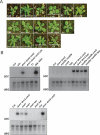
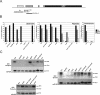


Comment in
-
Signaling silence--breaking ground and spreading out.Genes Dev. 2008 Jul 1;22(13):1719-23. doi: 10.1101/gad.1694608. Genes Dev. 2008. PMID: 18593872 Free PMC article.
References
-
- Alleman M., Sidorenko L., McGinnis K., Seshadri V., Dorweiler J.E., White J., Sikkink K., Chandler V.L. An RNA-dependent RNA polymerase is required for paramutation in maize. Nature. 2006;442:295–298. - PubMed
-
- Aufsatz W., Mette M.F., Matzke A.J., Matzke M. The role of MET1 in RNA-directed de novo and maintenance methylation of CG dinucleotides. Plant Mol. Biol. 2004;54:793–804. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
