Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites
- PMID: 18559960
- PMCID: PMC2483378
- DOI: 10.1105/tpc.108.058578
Arabidopsis ELONGATED MITOCHONDRIA1 is required for localization of DYNAMIN-RELATED PROTEIN3A to mitochondrial fission sites
Abstract
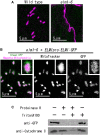
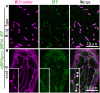
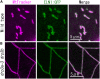
Mitochondrial fission is achieved partially by the activity of self-assembling dynamin-related proteins (DRPs) in diverse organisms. Mitochondrial fission in Arabidopsis thaliana is mediated by DRP3A and DRP3B, but the other genes and molecular mechanisms involved have yet to be elucidated. To identify these genes, we screened and analyzed Arabidopsis mutants with longer and fewer mitochondria than those of the wild type. ELM1 was found to be responsible for the phenotype of elongated mitochondria. This phenotype was also observed in drp3a plants. EST and genomic sequences similar to ELM1 were found in seed plants but not in other eukaryotes. ELM1:green fluorescent protein (GFP) was found to surround mitochondria, and ELM1 interacts with both DPR3A and DRP3B. In the elm1 mutant, DRP3A:GFP was observed in the cytosol, whereas in wild-type Arabidopsis, DRP3A:GFP localized to the ends and constricted sites of mitochondria. These results collectively suggest that mitochondrial fission in Arabidopsis is mediated by the plant-specific factor ELM1, which is required for the relocalization of DRP3A (and possibly also DRP3B) from the cytosol to mitochondrial fission sites.
Figures

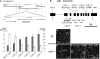




References
-
- Arimura, S., Aida, G.P., Fujimoto, M., Nakazono, M., and Tsutsumi, N. (2004. a). Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 45 236–242. - PubMed
-
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. - PubMed
-
- Bhar, D., Karren, M.A., Babst, M., and Shaw, J.M. (2006). Dimeric Dnm1-G385D interacts with mdv1 on mitochondria and can be stimulated to assemble into fission complexes containing Mdv1 and Fis1. J. Biol. Chem. 281 17312–17320. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

