Contemporary approaches for modifying the mouse genome
- PMID: 18559964
- PMCID: PMC2519963
- DOI: 10.1152/physiolgenomics.90242.2008
Contemporary approaches for modifying the mouse genome
Abstract
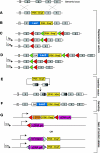
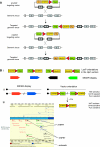
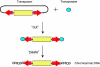
The mouse is a premiere experimental organism that has contributed significantly to our understanding of vertebrate biology. Manipulation of the mouse genome via embryonic stem (ES) cell technology makes it possible to engineer an almost limitless repertoire of mutations to model human disease and assess gene function. In this review we outline recent advances in mouse experimental genetics and provide a "how-to" guide for those people wishing to access this technology. We also discuss new technologies, such as transposon-mediated mutagenesis, and resources of targeting vectors and ES cells, which are likely to dramatically accelerate the pace with which we can assess gene function in vivo, and the progress of forward and reverse genetic screens in mice.
Figures


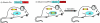
References
-
- Adams DJ, Biggs PJ, Cox T, Davies R, van der Weyden L, Jonkers J, Smith J, Plumb B, Taylor R, Nishijima I, Yu Y, Rogers J, Bradley A. Mutagenic insertion and chromosome engineering resource (MICER). Nat Genet 36: 867–871, 2004. - PubMed
-
- Adams DJ, Quail MA, Cox T, van der Weyden L, Gorick BD, Su Q, Chan WI, Davies R, Bonfield JK, Law F, Humphray S, Plumb B, Liu P, Rogers J, Bradley A. A genome-wide, end-sequenced 129Sv BAC library resource for targeting vector construction. Genomics 86: 753–758, 2005. - PubMed
-
- Austin S, Ziese M, Sternberg N. A novel role for site-specific recombination in maintenance of bacterial replicons. Cell 25: 729–736, 1981. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

