Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data
- PMID: 18560591
- PMCID: PMC2413010
- DOI: 10.1371/journal.pone.0002442
Estimation of isolation times of the island species in the Drosophila simulans complex from multilocus DNA sequence data
Abstract
Background: The Drosophila simulans species complex continues to serve as an important model system for the study of new species formation. The complex is comprised of the cosmopolitan species, D. simulans, and two island endemics, D. mauritiana and D. sechellia. A substantial amount of effort has gone into reconstructing the natural history of the complex, in part to infer the context in which functional divergence among the species has arisen. In this regard, a key parameter to be estimated is the initial isolation time (t) of each island species. Loci in regions of low recombination have lower divergence within the complex than do other loci, yet divergence from D. melanogaster is similar for both classes. This might reflect gene flow of the low-recombination loci subsequent to initial isolation, but it might also reflect differential effects of changing population size on the two recombination classes of loci when the low-recombination loci are subject to genetic hitchhiking or pseudohitchhiking
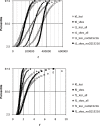
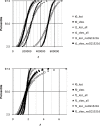
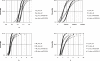
Methodology/principal findings: New DNA sequence variation data for 17 loci corroborate the prior observation from 13 loci that DNA sequence divergence is reduced in genes of low recombination. Two models are presented to estimate t and other relevant parameters (substitution rate correction factors in lineages leading to the island species and, in the case of the 4-parameter model, the ratio of ancestral to extant effective population size) from the multilocus DNA sequence data.
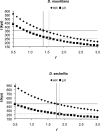
Conclusions/significance: In general, it appears that both island species were isolated at about the same time, here estimated at approximately 250,000 years ago. It also appears that the difference in divergence patterns of genes in regions of low and higher recombination can be reconciled by allowing a modestly larger effective population size for the ancestral population than for extant D. simulans.
Conflict of interest statement
Figures
References
-
- Throckmorton LH. The phylogeny, ecology and geography of Drosophila. In: King RC, editor. Handbook of Genetics, Volume 3, Invertebrates of Genetic Interest. New York: Plenum Publishing; 1975. pp. 421–470.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

