Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
- PMID: 18561946
- PMCID: PMC7094413
- DOI: 10.1016/j.jmb.2007.11.093
Solution structure of the c-terminal dimerization domain of SARS coronavirus nucleocapsid protein solved by the SAIL-NMR method
Abstract
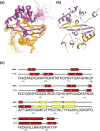
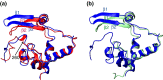
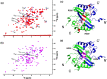
The C-terminal domain (CTD) of the severe acute respiratory syndrome coronavirus (SARS-CoV) nucleocapsid protein (NP) contains a potential RNA-binding region in its N-terminal portion and also serves as a dimerization domain by forming a homodimer with a molecular mass of 28 kDa. So far, the structure determination of the SARS-CoV NP CTD in solution has been impeded by the poor quality of NMR spectra, especially for aromatic resonances. We have recently developed the stereo-array isotope labeling (SAIL) method to overcome the size problem of NMR structure determination by utilizing a protein exclusively composed of stereo- and regio-specifically isotope-labeled amino acids. Here, we employed the SAIL method to determine the high-quality solution structure of the SARS-CoV NP CTD by NMR. The SAIL protein yielded less crowded and better resolved spectra than uniform (13)C and (15)N labeling, and enabled the homodimeric solution structure of this protein to be determined. The NMR structure is almost identical with the previously solved crystal structure, except for a disordered putative RNA-binding domain at the N-terminus. Studies of the chemical shift perturbations caused by the binding of single-stranded DNA and mutational analyses have identified the disordered region at the N-termini as the prime site for nucleic acid binding. In addition, residues in the beta-sheet region also showed significant perturbations. Mapping of the locations of these residues onto the helical model observed in the crystal revealed that these two regions are parts of the interior lining of the positively charged helical groove, supporting the hypothesis that the helical oligomer may form in solution.
Figures






Similar articles
-
Structure of the SARS coronavirus nucleocapsid protein RNA-binding dimerization domain suggests a mechanism for helical packaging of viral RNA.J Mol Biol. 2007 May 11;368(4):1075-86. doi: 10.1016/j.jmb.2007.02.069. Epub 2007 Mar 2. J Mol Biol. 2007. PMID: 17379242 Free PMC article.
-
Carboxyl terminus of severe acute respiratory syndrome coronavirus nucleocapsid protein: self-association analysis and nucleic acid binding characterization.Biochemistry. 2006 Oct 3;45(39):11827-35. doi: 10.1021/bi0609319. Biochemistry. 2006. PMID: 17002283
-
Multiple nucleic acid binding sites and intrinsic disorder of severe acute respiratory syndrome coronavirus nucleocapsid protein: implications for ribonucleocapsid protein packaging.J Virol. 2009 Mar;83(5):2255-64. doi: 10.1128/JVI.02001-08. Epub 2008 Dec 3. J Virol. 2009. PMID: 19052082 Free PMC article.
-
The SARS coronavirus nucleocapsid protein--forms and functions.Antiviral Res. 2014 Mar;103:39-50. doi: 10.1016/j.antiviral.2013.12.009. Epub 2014 Jan 11. Antiviral Res. 2014. PMID: 24418573 Free PMC article. Review.
-
[Progress in structural studies of larger proteins by the SAIL-NMR method].Tanpakushitsu Kakusan Koso. 2009 Sep;54(12 Suppl):1506-11. Tanpakushitsu Kakusan Koso. 2009. PMID: 21089580 Review. Japanese. No abstract available.
Cited by
-
SR/RS Motifs as Critical Determinants of Coronavirus Life Cycle.Front Mol Biosci. 2020 Aug 21;7:219. doi: 10.3389/fmolb.2020.00219. eCollection 2020. Front Mol Biosci. 2020. PMID: 32974389 Free PMC article.
-
Targeting the Interaction Between Spike Protein and Nucleocapsid Protein for Suppression and Detection of Human Coronavirus OC43.Front Immunol. 2022 Mar 10;13:835333. doi: 10.3389/fimmu.2022.835333. eCollection 2022. Front Immunol. 2022. PMID: 35359936 Free PMC article.
-
Non-encapsidation activities of the capsid proteins of positive-strand RNA viruses.Virology. 2013 Nov;446(1-2):123-32. doi: 10.1016/j.virol.2013.07.023. Epub 2013 Aug 27. Virology. 2013. PMID: 24074574 Free PMC article. Review.
-
Host DDX Helicases as Possible SARS-CoV-2 Proviral Factors: A Structural Overview of Their Hijacking Through Multiple Viral Proteins.Front Chem. 2020 Dec 10;8:602162. doi: 10.3389/fchem.2020.602162. eCollection 2020. Front Chem. 2020. PMID: 33381492 Free PMC article. Review.
-
SHAPE analysis of the RNA secondary structure of the Mouse Hepatitis Virus 5' untranslated region and N-terminal nsp1 coding sequences.Virology. 2015 Jan 15;475:15-27. doi: 10.1016/j.virol.2014.11.001. Epub 2014 Nov 21. Virology. 2015. PMID: 25462342 Free PMC article.
References
-
- Drosten C., Gunther S., Preiser W., van der Werf S., Brodt H.R., Becker S., et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T., Erdman D., Goldsmith C., Zaki S., Peret T., Emery S., et al. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
-
- Huang Q., Yu L., Petros A.M., Gunasekera A., Liu Z., Xu N., et al. Structure of the N-terminal RNA-binding domain of the SARS CoV nucleocapsid protein. Biochemistry. 2004;43:6059–6063. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

