A catalytic di-heme bis-Fe(IV) intermediate, alternative to an Fe(IV)=O porphyrin radical
- PMID: 18562294
- PMCID: PMC2438432
- DOI: 10.1073/pnas.0801643105
A catalytic di-heme bis-Fe(IV) intermediate, alternative to an Fe(IV)=O porphyrin radical
Abstract
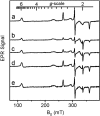
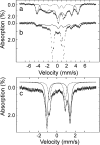
High-valent iron species are powerful oxidizing agents in chemical and biological catalysis. The best characterized form of an Fe(V) equivalent described in biological systems is the combination of a b-type heme with Fe(IV)=O and a porphyrin or amino acid cation radical (termed Compound I). This work describes an alternative natural mechanism to store two oxidizing equivalents above the ferric state for biological oxidation reactions. MauG is an enzyme that utilizes two covalently bound c-type hemes to catalyze the biosynthesis of the protein-derived cofactor tryptophan tryptophylquinone. Its natural substrate is a monohydroxylated tryptophan residue present in a 119-kDa precursor protein. An EPR-silent di-heme reaction intermediate of MauG was trapped. Mössbauer spectroscopy revealed the presence of two distinct Fe(IV) species. One is consistent with an Fe(IV)=O (ferryl) species (delta = 0.06 mm/s, DeltaE(Q) = 1.70 mm/s). The other is assigned to an Fe(IV) heme species with two axial ligands from protein (delta = 0.17 mm/s, DeltaE(Q) = 2.54 mm/s), which has never before been described in nature. This bis-Fe(IV) intermediate is remarkably stable but readily reacts with its native substrate. These findings broaden our views of how proteins can stabilize a highly reactive oxidizing species and the scope of enzyme-catalyzed posttranslational modifications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Tryptophan-mediated charge-resonance stabilization in the bis-Fe(IV) redox state of MauG.Proc Natl Acad Sci U S A. 2013 Jun 11;110(24):9639-44. doi: 10.1073/pnas.1301544110. Epub 2013 May 29. Proc Natl Acad Sci U S A. 2013. PMID: 23720312 Free PMC article.
-
Tryptophan tryptophylquinone biosynthesis: a radical approach to posttranslational modification.Biochim Biophys Acta. 2012 Nov;1824(11):1299-305. doi: 10.1016/j.bbapap.2012.01.008. Epub 2012 Jan 28. Biochim Biophys Acta. 2012. PMID: 22314272 Free PMC article. Review.
-
Heme iron nitrosyl complex of MauG reveals an efficient redox equilibrium between hemes with only one heme exclusively binding exogenous ligands.Biochemistry. 2009 Dec 15;48(49):11603-5. doi: 10.1021/bi9017544. Biochemistry. 2009. PMID: 19911786 Free PMC article.
-
Carboxyl group of Glu113 is required for stabilization of the diferrous and bis-Fe(IV) states of MauG.Biochemistry. 2013 Sep 17;52(37):6358-67. doi: 10.1021/bi400905s. Epub 2013 Aug 30. Biochemistry. 2013. PMID: 23952537 Free PMC article.
-
Uncovering novel biochemistry in the mechanism of tryptophan tryptophylquinone cofactor biosynthesis.Curr Opin Chem Biol. 2009 Oct;13(4):469-74. doi: 10.1016/j.cbpa.2009.06.026. Epub 2009 Aug 3. Curr Opin Chem Biol. 2009. PMID: 19648051 Free PMC article. Review.
Cited by
-
Characterization of the free energy dependence of an interprotein electron transfer reaction by variation of pH and site-directed mutagenesis.Biochim Biophys Acta. 2015 Oct;1847(10):1181-6. doi: 10.1016/j.bbabio.2015.06.012. Epub 2015 Jun 15. Biochim Biophys Acta. 2015. PMID: 26087387 Free PMC article.
-
Ascorbate protects the diheme enzyme, MauG, against self-inflicted oxidative damage by an unusual antioxidant mechanism.Biochem J. 2017 Jul 17;474(15):2563-2572. doi: 10.1042/BCJ20170349. Biochem J. 2017. PMID: 28634178 Free PMC article.
-
Escherichia coli Triheme Enzyme YhjA: Structure and Reactivity.Biochemistry. 2025 Aug 5;64(15):3322-3332. doi: 10.1021/acs.biochem.5c00202. Epub 2025 Jul 16. Biochemistry. 2025. PMID: 40669070
-
Electronic State of the His/Tyr-Ligated Heme of BthA by Mössbauer and DFT Analysis.Inorg Chem. 2020 Jul 20;59(14):10223-10233. doi: 10.1021/acs.inorgchem.0c01349. Epub 2020 Jun 30. Inorg Chem. 2020. PMID: 32602712 Free PMC article.
-
MauG, a diheme enzyme that catalyzes tryptophan tryptophylquinone biosynthesis by remote catalysis.Arch Biochem Biophys. 2014 Feb 15;544:112-8. doi: 10.1016/j.abb.2013.10.004. Epub 2013 Oct 19. Arch Biochem Biophys. 2014. PMID: 24144526 Free PMC article. Review.
References
-
- McIntire WS, Wemmer DE, Chistoserdov A, Lidstrom ME. A new cofactor in a prokaryotic enzyme: Tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991;252:817–824. - PubMed
-
- Davidson VL. Protein-derived cofactors. Expanding the scope of post-translational modifications. Biochemistry. 2007;46:5283–5292. - PubMed
-
- Davidson VL. Pyrroloquinoline quinone (PQQ) from methanol dehydrogenase and tryptophan tryptophylquinone (TTQ) from methylamine dehydrogenase. Adv Protein Chem. 2001;58:95–140. - PubMed
-
- Chen L, et al. Refined crystal structure of methylamine dehydrogenase from Paracoccus denitrificans at 1.75 Å resolution. J Mol Biol. 1998;276:131–149. - PubMed
-
- van der Palen CJ, et al. Mutational analysis of mau genes involved in methylamine metabolism in Paracoccus denitrificans. Eur J Biochem. 1995;230:860–871. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

