Guanine nucleotide exchange factor independence of the G-protein eEF1A through novel mutant forms and biochemical properties
- PMID: 18562321
- PMCID: PMC2517005
- DOI: 10.1074/jbc.M801095200
Guanine nucleotide exchange factor independence of the G-protein eEF1A through novel mutant forms and biochemical properties
Abstract
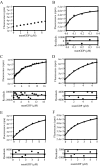
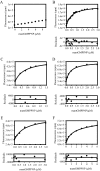
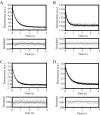
Most G-proteins require a guanine nucleotide exchange factor (GEF) to regulate a variety of critical cellular processes. Interestingly, a small number of G-proteins switch between the active and inactive forms without a GEF. Translation elongation factor 1A (eEF1A) normally requires the GEF eEF1Balpha to accelerate nucleotide dissociation. However, several mutant forms of eEF1A are functional independent of this essential regulator in vivo. GEF-independent eEF1A mutations localize close to the G-protein motifs that are crucial for nucleotide binding. Kinetic analysis demonstrated that reduced GDP affinity correlates with wild type growth and high translation activities of GEF-independent mutants. Furthermore, the mutant forms show an 11-22-fold increase in rates of GDP dissociation from eEF1A compared with the wild type protein. All mutant forms have dramatically enhanced stability at elevated temperatures. This, coupled with data demonstrating that eEF1A is also more stable in the presence of nucleotides, suggests that both the GEF and nucleotide have stabilizing effects on eEF1A. The biochemical properties of these eEF1A mutants provide insight into the mechanism behind GEF-independent G-protein function.
Figures


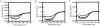

Similar articles
-
Mutations in a GTP-binding motif of eukaryotic elongation factor 1A reduce both translational fidelity and the requirement for nucleotide exchange.J Biol Chem. 1999 Oct 15;274(42):30297-302. doi: 10.1074/jbc.274.42.30297. J Biol Chem. 1999. PMID: 10514524
-
Unique classes of mutations in the Saccharomyces cerevisiae G-protein translation elongation factor 1A suppress the requirement for guanine nucleotide exchange.Genetics. 2006 Oct;174(2):651-63. doi: 10.1534/genetics.106.059899. Epub 2006 Sep 1. Genetics. 2006. PMID: 16951075 Free PMC article.
-
Coordination of eukaryotic translation elongation factor 1A (eEF1A) function in actin organization and translation elongation by the guanine nucleotide exchange factor eEF1Balpha.J Biol Chem. 2009 Feb 13;284(7):4739-47. doi: 10.1074/jbc.M807945200. Epub 2008 Dec 18. J Biol Chem. 2009. PMID: 19095653 Free PMC article.
-
The GAPs, GEFs, and GDIs of heterotrimeric G-protein alpha subunits.Int J Biol Sci. 2005;1(2):51-66. doi: 10.7150/ijbs.1.51. Epub 2005 Apr 1. Int J Biol Sci. 2005. PMID: 15951850 Free PMC article. Review.
-
eEF1A: thinking outside the ribosome.J Biol Chem. 2010 Jul 9;285(28):21209-13. doi: 10.1074/jbc.R110.113795. Epub 2010 May 5. J Biol Chem. 2010. PMID: 20444696 Free PMC article. Review.
Cited by
-
Translation elongation factor 1A mutants with altered actin bundling activity show reduced aminoacyl-tRNA binding and alter initiation via eIF2α phosphorylation.J Biol Chem. 2014 Jul 25;289(30):20928-38. doi: 10.1074/jbc.M114.570077. J Biol Chem. 2014. PMID: 24936063 Free PMC article.
-
Extensive proteomic remodeling is induced by eukaryotic translation elongation factor 1Bγ deletion in Aspergillus fumigatus.Protein Sci. 2013 Nov;22(11):1612-22. doi: 10.1002/pro.2367. Epub 2013 Sep 30. Protein Sci. 2013. PMID: 24023013 Free PMC article.
-
A Molecular Perspective on Mitochondrial Membrane Fusion: From the Key Players to Oligomerization and Tethering of Mitofusin.J Membr Biol. 2019 Oct;252(4-5):293-306. doi: 10.1007/s00232-019-00089-y. Epub 2019 Sep 4. J Membr Biol. 2019. PMID: 31485701 Review.
-
Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation.PLoS One. 2009 Jul 28;4(7):e6315. doi: 10.1371/journal.pone.0006315. PLoS One. 2009. PMID: 19636410 Free PMC article.
-
A novel protein domain induces high affinity selenocysteine insertion sequence binding and elongation factor recruitment.J Biol Chem. 2008 Dec 12;283(50):35129-39. doi: 10.1074/jbc.M806008200. Epub 2008 Oct 23. J Biol Chem. 2008. PMID: 18948268 Free PMC article.
References
-
- Vetter, I. R., and Wittinghofer, A. (2001) Science 294 1299-1304 - PubMed
-
- Sprang, S. R., and Coleman, D. E. (1998) Cell 95 155-158 - PubMed
-
- Bos, J. L., Rehmann, H., and Wittinghofer, A. (2007) Cell 129 865-877 - PubMed
-
- Pisareva, V. P., Hellen, C. U., and Pestova, T. V. (2007) Biochemistry 46 2622-2629 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

