Characterization and expression analysis of genes directing galactomannan synthesis in coffee
- PMID: 18562467
- PMCID: PMC2712370
- DOI: 10.1093/aob/mcn076
Characterization and expression analysis of genes directing galactomannan synthesis in coffee
Abstract
Background and aims: Galactomannans act as storage reserves for the seeds in some plants, such as guar (Cyamopsis tetragonoloba) and coffee (Coffea arabica and Coffea canephora). In coffee, the galactomannans can represent up to 25 % of the mass of the mature green coffee grain, and they exert a significant influence on the production of different types of coffee products. The objective of the current work was to isolate and characterize cDNA encoding proteins responsible for galactomannan synthesis in coffee and to study the expression of the corresponding transcripts in the developing coffee grain from C. arabica and C. canephora, which potentially exhibit slight galactomannan variations. Comparative gene expression analysis was also carried out for several other tissues of C. arabica and C. canephora.
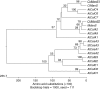
Methods: cDNA banks, RACE-PCR and genome walking were used to generate full-length cDNA for two putative coffee mannan synthases (ManS) and two galactomannan galactosyl transferases (GMGT). Gene-specific probe-primer sets were then generated and used to carry out comparative expression analysis of the corresponding genes in different coffee tissues using quantitative RT-PCR.
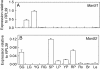
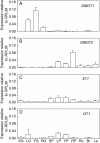
Key results: Two of the putative galactomannan biosynthetic genes, ManS1 and GMGT1, were demonstrated to have very high expression in the developing coffee grain of both Coffea species during endosperm development, consistent with our proposal that these two genes are responsible for the production of the majority of the galactomannans found in the grain. In contrast, the expression data presented indicates that the ManS2 gene product is probably involved in the synthesis of the galactomannans found in green tissue.
Conclusions: The identification of genes implicated in galactomannan synthesis in coffee are presented. The data obtained will enable more detailed studies on the biosynthesis of this important component of coffee grain and contribute to a better understanding of some functional differences between grain from C. arabica and C. canephora.
Figures

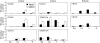

References
-
- Bailey RW. Polysaccharides in the leguminosae. In: Harborne JB, Boulter D, Turner BL, editors. Chemotaxonomy of the Leguminosae. London: Academic Press; 1971. pp. 503–541.
-
- Bradbury AGW, Halliday DJ. Chemical structures of green coffee bean polysaccharides. Journal of Agricultural and Food Chemistry. 1990;38:389–392.
-
- Cavalier DM, Keegstra K. Two xyloglucan xylosyltransferases catalyze the addition of multiple xylosyl residues to cellohexaose. Journal of Biological Chemistry. 2006;281:34197–34207. - PubMed
-
- Clifford MN. Chemical and physical aspects of green coffee and coffee products. In: Clifford MN, Willson KC, editors. Coffee, botany, biochemistry and production of beans and beverage. London: Croom Helm; 1985. pp. 305–374.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

