The temporal dynamics of processes underlying Y chromosome degeneration
- PMID: 18562655
- PMCID: PMC2475751
- DOI: 10.1534/genetics.107.084012
The temporal dynamics of processes underlying Y chromosome degeneration
Abstract
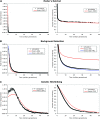
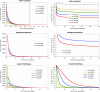
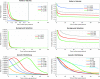
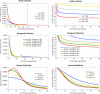
Y chromosomes originate from ordinary autosomes and degenerate by accumulating deleterious mutations. This accumulation results from a lack of recombination on the Y and is driven by interference among deleterious mutations (Muller's ratchet and background selection) and the fixation of beneficial alleles (genetic hitchhiking). Here I show that the relative importance of these processes is expected to vary over the course of Y chromosome evolution due to changes in the number of active genes. The dominant mode of degeneration on a newly formed gene-rich Y chromosome is expected to be Muller's ratchet and/or background selection due to the large numbers of deleterious mutations arising in active genes. However, the relative importance of these modes of degeneration declines rapidly as active genes are lost. In contrast, the rate of degeneration due to hitchhiking is predicted to be highest on Y chromosomes containing an intermediate number of active genes. The temporal dynamics of these processes imply that a gradual restriction of recombination, as inferred in mammals, will increase the importance of genetic hitchhiking relative to Muller's ratchet and background selection.
Figures




References
-
- Aitken, R., and J. Marshall Graves, 2002. The future of sex. Nature 415 963. - PubMed
-
- Bachtrog, D., 2004. Evidence that positive selection drives Y-chromosome degeneration in Drosophila miranda. Nat. Genet. 36 518–522. - PubMed
-
- Bachtrog, D., and I. Gordo, 2004. Adaptive evolution of asexual populations under Muller's ratchet. Evol. Int. J. Org. Evol. 58 1403–1413. - PubMed
-
- Barton, N. H., and B. Charlesworth, 1998. Why sex and recombination? Science 281 1986–1990. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

