Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae
- PMID: 18562657
- PMCID: PMC2475728
- DOI: 10.1534/genetics.107.083493
Meiotic recombination at the ends of chromosomes in Saccharomyces cerevisiae
Abstract
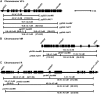
Meiotic reciprocal recombination (crossing over) was examined in the outermost 60-80 kb of almost all Saccharomyces cerevisiae chromosomes. These sequences included both repetitive gene-poor subtelomeric heterochromatin-like regions and their adjacent unique gene-rich euchromatin-like regions. Subtelomeric sequences underwent very little crossing over, exhibiting approximately two- to threefold fewer crossovers per kilobase of DNA than the genomic average. Surprisingly, the adjacent euchromatic regions underwent crossing over at twice the average genomic rate and contained at least nine new recombination "hot spots." These results prompted an analysis of existing genetic mapping data, which showed that meiotic reciprocal recombination rates were on average greater near chromosome ends exclusive of the subtelomeres. Thus, the distribution of crossovers in S. cerevisiae appears to resemble that found in several higher eukaryotes where the outermost chromosomal regions show increased crossing over.
Figures





References
-
- Barlow, A. L., and M. A. Hulten, 1998. Crossing over analysis at pachytene in man. Eur. J. Hum. Genet. 6 350–358. - PubMed
-
- Blitzblau, H. G., G. W. Bell, J. Rodriquez, S. P. Bell and A. Hochwagen, 2007. Mapping of meiotic single-stranded DNA reveals double-strand break hotspots near telomeres and centromeres. Curr. Biol. 17 2003–2012. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

