The fission yeast BLM homolog Rqh1 promotes meiotic recombination
- PMID: 18562672
- PMCID: PMC2475723
- DOI: 10.1534/genetics.108.088955
The fission yeast BLM homolog Rqh1 promotes meiotic recombination
Abstract
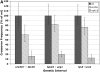
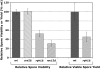
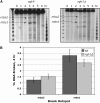
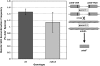
RecQ helicases are found in organisms as diverse as bacteria, fungi, and mammals. These proteins promote genome stability, and mutations affecting human RecQ proteins underlie premature aging and cancer predisposition syndromes, including Bloom syndrome, caused by mutations affecting the BLM protein. In this study we show that mutants lacking the Rqh1 protein of the fission yeast Schizosaccharomyces pombe, a RecQ and BLM homolog, have substantially reduced meiotic recombination, both gene conversions and crossovers. The relative proportion of gene conversions having associated crossovers is unchanged from that in wild type. In rqh1 mutants, meiotic DNA double-strand breaks are formed and disappear with wild-type frequency and kinetics, and spore viability is only moderately reduced. Genetic analyses and the wild-type frequency of both intersister and interhomolog joint molecules argue against these phenotypes being explained by an increase in intersister recombination at the expense of interhomolog recombination. We suggest that Rqh1 extends hybrid DNA and biases the recombination outcome toward crossing over. Our results contrast dramatically with those from the budding yeast ortholog, Sgs1, which has a meiotic antirecombination function that suppresses recombination events involving more than two DNA duplexes. These observations underscore the multiple recombination functions of RecQ homologs and emphasize that even conserved proteins can be adapted to play different roles in different organisms.
Figures



Similar articles
-
Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases.Genes (Basel). 2020 Feb 18;11(2):205. doi: 10.3390/genes11020205. Genes (Basel). 2020. PMID: 32085395 Free PMC article. Review.
-
Yeast as a model system to study RecQ helicase function.DNA Repair (Amst). 2010 Mar 2;9(3):303-14. doi: 10.1016/j.dnarep.2009.12.007. Epub 2010 Jan 13. DNA Repair (Amst). 2010. PMID: 20071248 Review.
-
The Rad52 homologs Rad22 and Rti1 of Schizosaccharomyces pombe are not essential for meiotic interhomolog recombination, but are required for meiotic intrachromosomal recombination and mating-type-related DNA repair.Genetics. 2008 Apr;178(4):2399-412. doi: 10.1534/genetics.107.085696. Genetics. 2008. PMID: 18430957 Free PMC article.
-
Mus81-Eme1-dependent and -independent crossovers form in mitotic cells during double-strand break repair in Schizosaccharomyces pombe.Mol Cell Biol. 2007 May;27(10):3828-38. doi: 10.1128/MCB.01596-06. Epub 2007 Mar 12. Mol Cell Biol. 2007. PMID: 17353272 Free PMC article.
-
Genetic interactions between the chromosome axis-associated protein Hop1 and homologous recombination determinants in Schizosaccharomyces pombe.Curr Genet. 2018 Oct;64(5):1089-1104. doi: 10.1007/s00294-018-0827-7. Epub 2018 Mar 17. Curr Genet. 2018. PMID: 29550859 Free PMC article.
Cited by
-
Rad51/Dmc1 paralogs and mediators oppose DNA helicases to limit hybrid DNA formation and promote crossovers during meiotic recombination.Nucleic Acids Res. 2014 Dec 16;42(22):13723-35. doi: 10.1093/nar/gku1219. Epub 2014 Nov 20. Nucleic Acids Res. 2014. PMID: 25414342 Free PMC article.
-
Mammalian BLM helicase is critical for integrating multiple pathways of meiotic recombination.J Cell Biol. 2010 Mar 22;188(6):779-89. doi: 10.1083/jcb.200909048. J Cell Biol. 2010. PMID: 20308424 Free PMC article.
-
The RecQ DNA helicases in DNA repair.Annu Rev Genet. 2010;44:393-417. doi: 10.1146/annurev-genet-102209-163602. Annu Rev Genet. 2010. PMID: 21047263 Free PMC article. Review.
-
Maintenance of Yeast Genome Integrity by RecQ Family DNA Helicases.Genes (Basel). 2020 Feb 18;11(2):205. doi: 10.3390/genes11020205. Genes (Basel). 2020. PMID: 32085395 Free PMC article. Review.
-
A failure of meiotic chromosome segregation in a fbh1Delta mutant correlates with persistent Rad51-DNA associations.Nucleic Acids Res. 2011 Mar;39(5):1718-31. doi: 10.1093/nar/gkq977. Epub 2010 Dec 11. Nucleic Acids Res. 2011. PMID: 21149262 Free PMC article.
References
-
- Ahmad, F., and E. Stewart, 2005. The N-terminal region of the Schizosaccharomyces pombe RecQ helicase, Rqh1p, physically interacts with Topoisomerase III and is required for Rqh1p function. Mol. Genet. Genomics 273 102–114. - PubMed
-
- Bennett, R. J., J. L. Keck and J. C. Wang, 1999. Binding specificity determines polarity of DNA unwinding by the Sgs1 protein of S. cerevisiae. J. Mol. Biol. 289 235–248. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

