Polyubiquitylation of histone H2B
- PMID: 18562693
- PMCID: PMC2526708
- DOI: 10.1091/mbc.e08-01-0050
Polyubiquitylation of histone H2B
Abstract
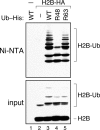
Covalent modification of histones by ubiquitylation is a prominent epigenetic mark that features in a variety of chromatin-based events such as histone methylation, gene silencing, and repair of DNA damage. The prototypical example of histone ubiquitylation is that of histone H2B in Saccharomyces cerevisiae. In this case, attachment of ubiquitin to lysine 123 (K123) of H2B is important for regulation of both active and transcriptionally silent genes and participates in trans to signal methylation of histone H3. It is generally assumed that H2B is monoubiquitylated at K123 and that it is this single ubiquitin moiety that influences H2B function. To determine whether this assumption is correct, we have re-examined the ubiquitylation status of endogenous H2B in yeast. We find that, contrary to expectations, H2B is extensively polyubiquitylated. Polyubiquitylation of H2B appears to occur within the context of chromatin and is not associated with H2B destruction. There are at least two distinct modes of H2B polyubiquitylation: one that occurs at K123 and depends on the Rad6-Bre1 ubiquitylation machinery and another that occurs on multiple lysine residues and is catalyzed by an uncharacterized ubiquitin ligase(s). Interestingly, these ubiquitylation events are under the influence of different combinations of ubiquitin-specific proteases, suggesting that they have distinct biological functions. These results raise the possibility that some of the biological effects of ubiquitylation of H2B are exerted via ubiquitin chains, rather than a single ubiquitin group.
Figures
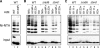



References
-
- Bloom J., Amador V., Bartolini F., DeMartino G., Pagano M. Proteasome-mediated degradation of p21 via N-terminal ubiquitinylation. Cell. 2003;115:71–82. - PubMed
-
- Crook T., Ludwig R. L., Marston N. J., Willkomm D., Vousden K. H. Sensitivity of p53 lysine mutants to ubiquitin-directed degradation targeted by human papillomavirus E6. Virology. 1996;217:285–292. - PubMed
-
- Daniel J. A., Torok M. S., Sun Z. W., Schieltz D., Allis C. D., Yates J. R., 3rd, Grant P. A. Deubiquitination of histone H2B by a yeast acetyltransferase complex regulates transcription. J. Biol. Chem. 2004;279:1867–1871. - PubMed
-
- Davie J. R., Delcuve G. P., Nickel B. E., Moirier R., Bailey G. Reduced levels of histones H1o and H1b, and unaltered content of methylated DNA in rainbow trout hepatocellular carcinoma chromatin. Cancer Res. 1987;47:5407–5410. - PubMed
-
- Davie J. R., Lin R., Allis C. D. Timing of the appearance of ubiquitinated histones in developing new macronuclei of Tetrahymena thermophila. Biochem. Cell Biol. 1991;69:66–71. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

