Assembly reflects evolution of protein complexes
- PMID: 18563089
- PMCID: PMC2658002
- DOI: 10.1038/nature06942
Assembly reflects evolution of protein complexes
Abstract
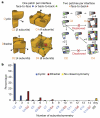
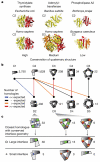
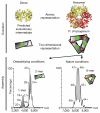
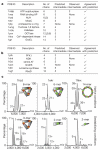
A homomer is formed by self-interacting copies of a protein unit. This is functionally important, as in allostery, and structurally crucial because mis-assembly of homomers is implicated in disease. Homomers are widespread, with 50-70% of proteins with a known quaternary state assembling into such structures. Despite their prevalence, their role in the evolution of cellular machinery and the potential for their use in the design of new molecular machines, little is known about the mechanisms that drive formation of homomers at the level of evolution and assembly in the cell. Here we present an analysis of over 5,000 unique atomic structures and show that the quaternary structure of homomers is conserved in over 70% of protein pairs sharing as little as 30% sequence identity. Where quaternary structure is not conserved among the members of a protein family, a detailed investigation revealed well-defined evolutionary pathways by which proteins transit between different quaternary structure types. Furthermore, we show by perturbing subunit interfaces within complexes and by mass spectrometry analysis, that the (dis)assembly pathway mimics the evolutionary pathway. These data represent a molecular analogy to Haeckel's evolutionary paradigm of embryonic development, where an intermediate in the assembly of a complex represents a form that appeared in its own evolutionary history. Our model of self-assembly allows reliable prediction of evolution and assembly of a complex solely from its crystal structure.
Figures
References
-
- Cabezon E, et al. Homologous and heterologous inhibitory effects of ATPase inhibitor proteins on F-ATPases. J. Biol. Chem. 2002;277:41334–41341. - PubMed
-
- Hardy LW, et al. Atomic structure of thymidylate synthase: target for rational drug design. Science. 1987;235:448–455. - PubMed
-
- Iber D, Clarkson J, Yudkin MD, Campbell ID. The mechanism of cell differentiation in Bacillus subtilis. Nature. 2006;441:371–374. - PubMed
-
- Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem. Sci. 2004;29:618–625. - PubMed
-
- Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J. Mol. Biol. 1965;12:88–118. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

