Position of human chromosomes is conserved in mouse nuclei indicating a species-independent mechanism for maintaining genome organization
- PMID: 18563425
- PMCID: PMC2614925
- DOI: 10.1007/s00412-008-0171-7
Position of human chromosomes is conserved in mouse nuclei indicating a species-independent mechanism for maintaining genome organization
Abstract
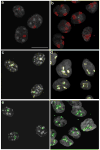
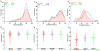
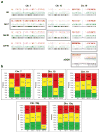
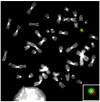
The nonrandom positioning of chromosome territories in eukaryotic cells is largely correlated with gene density and is conserved throughout evolution. Gene-rich chromosomes are predominantly central, while gene-poor chromosomes are peripherally localized in interphase nuclei. We previously demonstrated that artificially introduced human chromosomes assume a position equivalent to their endogenous homologues in the diploid colon cancer cell line DLD-1. These chromosomal aneuploidies result in a significant increase in transcript levels, suggesting a relationship between genomic copy number, gene expression, and chromosome position. We previously proposed that each chromosome is marked by a "zip code" that determines its nonrandom position in the nucleus. In this paper, we investigated (1) whether mouse nuclei recognize such determinants of nuclear position on human chromosomes to facilitate their distinct partitioning and (2) if chromosome positioning and transcriptional activity remain coupled under these trans-species conditions. Using three-dimensional fluorescence in situ hybridization, confocal microscopy, and gene expression profiling, we show (1) that gene-poor and gene-rich human chromosomes maintain their divergent but conserved positions in mouse-human hybrid nuclei and (2) that a foreign human chromosome is actively transcribed in mouse nuclei. Our results suggest a species-independent conserved mechanism for the nonrandom positioning of chromosomes in the three-dimensional interphase nucleus.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

