Genomic structure and expression of Jmjd6 and evolutionary analysis in the context of related JmjC domain containing proteins
- PMID: 18564434
- PMCID: PMC2453528
- DOI: 10.1186/1471-2164-9-293
Genomic structure and expression of Jmjd6 and evolutionary analysis in the context of related JmjC domain containing proteins
Abstract
Background: The jumonji C (JmjC) domain containing gene 6 (Jmjd6, previously known as phosphatidylserine receptor) has misleadingly been annotated to encode a transmembrane receptor for the engulfment of apoptotic cells. Given the importance of JmjC domain containing proteins in controlling a wide range of diverse biological functions, we undertook a comparative genomic analysis to gain further insights in Jmjd6 gene organisation, evolution, and protein function.
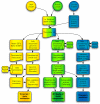
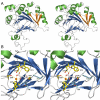
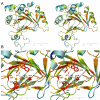
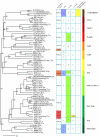
Results: We describe here a semiautomated computational pipeline to identify and annotate JmjC domain containing proteins. Using a sequence segment N-terminal of the Jmjd6 JmjC domain as query for a reciprocal BLAST search, we identified homologous sequences in 62 species across all major phyla. Retrieved Jmjd6 sequences were used to phylogenetically analyse corresponding loci and their genomic neighbourhood. This analysis let to the identification and characterisation of a bi-directional transcriptional unit compromising the Jmjd6 and 1110005A03Rik genes and to the recognition of a new, before overseen Jmjd6 exon in mammals. Using expression studies, two novel Jmjd6 splice variants were identified and validated in vivo. Analysis of the Jmjd6 neighbouring gene 1110005A03Rik revealed an incident deletion of this gene in two out of three earlier reported Jmjd6 knockout mice, which might affect previously described conflicting phenotypes. To determine potentially important residues for Jmjd6 function a structural model of the Jmjd6 protein was calculated based on sequence conservation. This approach identified a conserved double-stranded beta-helix (DSBH) fold and a HxDxnH facial triad as structural motifs. Moreover, our systematic annotation in nine species identified 313 DSBH fold-containing proteins that split into 25 highly conserved subgroups.
Conclusion: We give further evidence that Jmjd6 most likely has a function as a nonheme-Fe(II)-2-oxoglutarate-dependent dioxygenase as previously suggested. Further, we provide novel insights into the evolution of Jmjd6 and other related members of the superfamily of JmjC domain containing proteins. Finally, we discuss possibilities of the involvement of Jmjd6 and 1110005A03Rik in an antagonistic biochemical pathway.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

