Human embryonic stem cells have enhanced repair of multiple forms of DNA damage
- PMID: 18566332
- PMCID: PMC2574957
- DOI: 10.1634/stemcells.2007-1041
Human embryonic stem cells have enhanced repair of multiple forms of DNA damage
Abstract
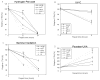
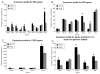
Embryonic stem cells need to maintain genomic integrity so that they can retain the ability to differentiate into multiple cell types without propagating DNA errors. Previous studies have suggested that mechanisms of genome surveillance, including DNA repair, are superior in mouse embryonic stem cells compared with various differentiated murine cells. Using single-cell gel electrophoresis (comet assay) we found that human embryonic stem cells (BG01, I6) have more efficient repair of different types of DNA damage (generated from H2O2, UV-C, ionizing radiation, or psoralen) than human primary fibroblasts (WI-38, hs27) and, with the exception of UV-C damage, HeLa cells. Microarray gene expression analysis showed that mRNA levels of several DNA repair genes are elevated in human embryonic stem cells compared with their differentiated forms (embryoid bodies). These data suggest that genomic maintenance pathways are enhanced in human embryonic stem cells, relative to differentiated human cells.
Figures




References
-
- Bohr VA. Repair of oxidative DNA damage in nuclear and mitochondrial DNA, and some changes with aging in mammalian cells. Free Radic Biol Med. 2002;32:804–812. - PubMed
-
- Houtgraaf JH, Versmissen J, van der Giessen WJ. A concise review of DNA damage checkpoints and repair in mammalian cells. Cardiovasc Revasc Med. 2006;7:165–172. - PubMed
-
- Peterson CL, Cote J. Cellular machineries for chromosomal DNA repair. Genes Dev. 2004;18:602–616. - PubMed
-
- Sancar A, Lindsey-Boltz LA, Unsal-Kacmaz K, Linn S. Molecular mechanisms of mammalian DNA repair and the DNA damage checkpoints. Annual Review of Biochemistry. 2004;73:39–85. - PubMed
-
- Andressoo JO, Hoeijmakers JH, Mitchell JR. Nucleotide excision repair disorders and the balance between cancer and aging. Cell Cycle. 2006;5:2886–2888. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

