Polyglutamylation: a fine-regulator of protein function? 'Protein Modifications: beyond the usual suspects' review series
- PMID: 18566597
- PMCID: PMC2475320
- DOI: 10.1038/embor.2008.114
Polyglutamylation: a fine-regulator of protein function? 'Protein Modifications: beyond the usual suspects' review series
Abstract
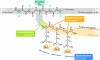
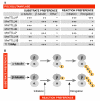
Polyglutamylation is a post-translational modification in which glutamate side chains of variable lengths are formed on the modified protein. It is evolutionarily conserved from protists to mammals and its most prominent substrate is tubulin, the microtubule (MT) building block. Various polyglutamylation states of MTs can be distinguished within a single cell and they are also characteristic of specific cell types or organelles. Polyglutamylation has been proposed to be involved in the functional adaptation of MTs, as it occurs within the carboxy-terminal tubulin tails that participate directly in the binding of many structural and motor MT-associated proteins. The discovery of a new family of enzymes that catalyse this modification has brought new insight into the mechanism of polyglutamylation and now allows for direct functional studies of the role of tubulin polyglutamylation. Moreover, the recent identification of new substrates of polyglutamylation indicates that this post-translational modification could be a potential regulator of diverse cellular processes.
Figures



References
-
- Amos LA (2000) Focusing-in on microtubules. Curr Opin Struct Biol 10: 236–241 - PubMed
-
- Audebert S, Koulakoff A, Berwald-Netter Y, Gros F, Denoulet P, Edde B (1994) Developmental regulation of polyglutamylated α- and β-tubulin in mouse brain neurons. J Cell Sci 107: 2313–2322 - PubMed
-
- Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M (1998a) Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton 39: 223–232 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

