QM/QM studies for Michael reaction in coronavirus main protease (3CL Pro)
- PMID: 18567519
- PMCID: PMC7110475
- DOI: 10.1016/j.jmgm.2008.05.002
QM/QM studies for Michael reaction in coronavirus main protease (3CL Pro)
Abstract
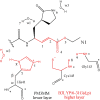
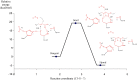
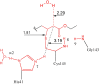
Severe acute respiratory syndrome (SARS) is an illness caused by a novel corona virus wherein the main proteinase called 3CL(Pro) has been established as a target for drug design. The mechanism of action involves nucleophilic attack by Cys145 present in the active site on the carbonyl carbon of the scissile amide bond of the substrate and the intermediate product is stabilized by hydrogen bonds with the residues of the oxyanion hole. Based on the X-ray structure of 3CL(Pro) co-crystallized with a trans-alpha,beta-unsaturated ethyl ester (Michael acceptor), a set of QM/QM and QM/MM calculations were performed, yielding three models with increasingly higher the number of atoms. A previous validation step was performed using classical theoretical calculation and PROCHECK software. The Michael reaction studies show an exothermic process with -4.5 kcal/mol. During the reaction pathway, an intermediate is formed by hydrogen and water molecule migration from a histidine residue and solvent, respectively. In addition, similar with experimental results, the complex between N3 and 3CL(Pro) is 578 kcal/mol more stable than N1-3CL(Pro) using Own N-layer Integrated molecular Orbital molecular Mechanics (ONIOM) approach. We suggest 3CL(Pro) inhibitors need small polar groups to decrease the energy barrier for alkylation reaction. These results can be useful for the development of new compounds against SARS.
Figures



Similar articles
-
A mechanistic view of enzyme inhibition and peptide hydrolysis in the active site of the SARS-CoV 3C-like peptidase.J Mol Biol. 2007 Aug 24;371(4):1060-74. doi: 10.1016/j.jmb.2007.06.001. Epub 2007 Jun 8. J Mol Biol. 2007. PMID: 17599357 Free PMC article.
-
Maturation mechanism of severe acute respiratory syndrome (SARS) coronavirus 3C-like proteinase.J Biol Chem. 2010 Sep 3;285(36):28134-40. doi: 10.1074/jbc.M109.095851. Epub 2010 May 20. J Biol Chem. 2010. PMID: 20489209 Free PMC article.
-
Design, synthesis, and evaluation of trifluoromethyl ketones as inhibitors of SARS-CoV 3CL protease.Bioorg Med Chem. 2008 Apr 15;16(8):4652-60. doi: 10.1016/j.bmc.2008.02.040. Epub 2008 Feb 15. Bioorg Med Chem. 2008. PMID: 18329272 Free PMC article.
-
Design and Evaluation of Anti-SARS-Coronavirus Agents Based on Molecular Interactions with the Viral Protease.Molecules. 2020 Aug 27;25(17):3920. doi: 10.3390/molecules25173920. Molecules. 2020. PMID: 32867349 Free PMC article. Review.
-
Drug design targeting the main protease, the Achilles' heel of coronaviruses.Curr Pharm Des. 2006;12(35):4573-90. doi: 10.2174/138161206779010369. Curr Pharm Des. 2006. PMID: 17168763 Review.
Cited by
-
Crystal structure of the papain-like protease of MERS coronavirus reveals unusual, potentially druggable active-site features.Antiviral Res. 2014 Sep;109:72-82. doi: 10.1016/j.antiviral.2014.06.011. Epub 2014 Jun 30. Antiviral Res. 2014. PMID: 24992731 Free PMC article.
-
Mechanisms of Proteolytic Enzymes and Their Inhibition in QM/MM Studies.Int J Mol Sci. 2021 Mar 22;22(6):3232. doi: 10.3390/ijms22063232. Int J Mol Sci. 2021. PMID: 33810118 Free PMC article. Review.
-
Room-temperature X-ray crystallography reveals the oxidation and reactivity of cysteine residues in SARS-CoV-2 3CL Mpro: insights into enzyme mechanism and drug design.IUCrJ. 2020 Sep 21;7(Pt 6):1028-35. doi: 10.1107/S2052252520012634. Online ahead of print. IUCrJ. 2020. PMID: 33063790 Free PMC article.
-
Antimalarial activity of 4-metoxychalcones: docking studies as falcipain/plasmepsin inhibitors, ADMET and lipophilic efficiency analysis to identify a putative oral lead candidate.Molecules. 2013 Dec 10;18(12):15276-87. doi: 10.3390/molecules181215276. Molecules. 2013. PMID: 24335577 Free PMC article.
-
Supercomputer simulation of the covalent inhibition of the main protease of SARS-CoV-2.Russ Chem Bull. 2021;70(11):2084-2089. doi: 10.1007/s11172-021-3319-8. Epub 2022 Jan 15. Russ Chem Bull. 2021. PMID: 35068913 Free PMC article.
References
-
- Lee N., Hui D., Wu A., Chan P., Cameron P., Joynt G.M. A major outbreak of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 2003;348:1986–1994. - PubMed
-
- Drosten C., Günther S., Preiser W., van der Werf S., Brodt H., Becker S., Rabenau H., Panning M., Kolesnikova L., Fouchier R.A.M., Berger A., Burguière A.M., Cinatl J., Eickmann M., Escriou N., Grywna K., Kramme S., Manuguerra J.C., Müller S., Rickerts V., Stürmer M., Vieth S., Klenk H., Osterhaus A.D.M.E., Schmitz H., Doerr H.W. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 2003;348:1967–1976. - PubMed
-
- Ksiazek T.G., Erdman D., Goldsmith C.S., Zaki S.R., Peret T., Emery S., Tong S., Urbani C., Comer J.A., Lim W., Rollin P.E., Dowell S.F., Ling A., Humphrey C.D., Shieh W., Guarner J., Paddock C.D., Rota P., Fields B., DeRisi J., Yang J., Cox N., Hughes J.M., LeDuc J.W., Bellini W.J., Anderson L.J. Enteric involvement of severe acute respiratory syndrome-associated coronavirus infection. N. Engl. J. Med. 2003;348:1953–1966. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

