Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5' untranslated regions are efficiently translated in cells by a cap-dependent mechanism
- PMID: 18567818
- PMCID: PMC2491466
- DOI: 10.1261/rna.1039708
Monocistronic mRNAs containing defective hepatitis C virus-like picornavirus internal ribosome entry site elements in their 5' untranslated regions are efficiently translated in cells by a cap-dependent mechanism
Abstract
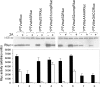
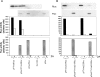
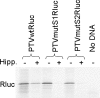
The initiation of protein synthesis on mRNAs within eukaryotic cells is achieved either by a 5' cap-dependent mechanism or through internal initiation directed by an internal ribosome entry site (IRES). Picornavirus IRES elements, located in the 5' untranslated region (5'UTR), contain extensive secondary structure and multiple upstream AUG codons. These features can be expected to inhibit cap-dependent initiation of translation. However, we have now shown that certain mutant hepatitis C virus-like picornavirus IRES elements (from porcine teschovirus-1 and avian encephalomyelitis virus), which are unable to direct internal initiation, are not significant barriers to efficient translation of capped monocistronic mRNAs that contain these defective elements within their 5'UTRs. Moreover, the translation of these mRNAs is highly sensitive to the expression of an enterovirus 2A protease (which induces cleavage of eIF4G) and is also inhibited by hippuristanol, a specific inhibitor of eIF4A function, in contrast to their parental wild-type IRES elements. These results provide a possible basis for the evolution of viral IRES elements within the context of functional mRNAs that are translated by a cap-dependent mechanism.
Figures



References
-
- Belsham G.J., Jackson R.J. Translation initiation on picornavirus RNA. In: Sonenberg N., editor. Translational Control of Gene Expression. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2000. pp. 869–900. Cold Spring Harbor Monograph 39,
-
- Bordeleau M.E., Mori A., Oberer M., Lindqvist L., Chard L.S., Higa T., Belsham G.J., Wagner G., Tanaka J., Pelletier J. Functional characterization of IRESes by an inhibitor of the RNA helicase eIF4A. Nature Chem. Biol. 2006;2:213–220. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
