Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme
- PMID: 18568847
- PMCID: PMC2649671
- DOI: 10.1080/10409230802058320
Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme
Erratum in
- Crit Rev Biochem Mol Biol. 2008 Jul-Aug;43(4):287-8
Abstract
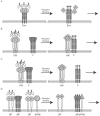
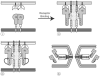
Recent work has identified three distinct classes of viral membrane fusion proteins based on structural criteria. In addition, there are at least four distinct mechanisms by which viral fusion proteins can be triggered to undergo fusion-inducing conformational changes. Viral fusion proteins also contain different types of fusion peptides and vary in their reliance on accessory proteins. These differing features combine to yield a rich diversity of fusion proteins. Yet despite this staggering diversity, all characterized viral fusion proteins convert from a fusion-competent state (dimers or trimers, depending on the class) to a membrane-embedded homotrimeric prehairpin, and then to a trimer-of-hairpins that brings the fusion peptide, attached to the target membrane, and the transmembrane domain, attached to the viral membrane, into close proximity thereby facilitating the union of viral and target membranes. During these conformational conversions, the fusion proteins induce membranes to progress through stages of close apposition, hemifusion, and then the formation of small, and finally large, fusion pores. Clearly, highly divergent proteins have converged on the same overall strategy to mediate fusion, an essential step in the life cycle of every enveloped virus.
Figures


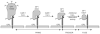

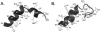
References
-
- Aguilar HC, Matreyek KA, Filone CM, Hashimi ST, Levroney EL, Negrete OA, Bertolotti-Ciarlet A, Choi DY, McHardy I, Fulcher JA, Su SV, Wolf MC, Kohatsu L, Baum LG, Lee B. N-Glycans on Nipah virus fusion protein protect against neutralization but reduce membrane fusion and viral entry. J Virol. 2006;80:4878–4889. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
