Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species
- PMID: 18570682
- PMCID: PMC2443145
- DOI: 10.1186/1471-2229-8-70
Complete nucleotide sequence of the Cryptomeria japonica D. Don. chloroplast genome and comparative chloroplast genomics: diversified genomic structure of coniferous species
Abstract
Background: The recent determination of complete chloroplast (cp) genomic sequences of various plant species has enabled numerous comparative analyses as well as advances in plant and genome evolutionary studies. In angiosperms, the complete cp genome sequences of about 70 species have been determined, whereas those of only three gymnosperm species, Cycas taitungensis, Pinus thunbergii, and Pinus koraiensis have been established. The lack of information regarding the gene content and genomic structure of gymnosperm cp genomes may severely hamper further progress of plant and cp genome evolutionary studies. To address this need, we report here the complete nucleotide sequence of the cp genome of Cryptomeria japonica, the first in the Cupressaceae sensu lato of gymnosperms, and provide a comparative analysis of their gene content and genomic structure that illustrates the unique genomic features of gymnosperms.
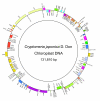
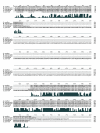
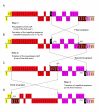
Results: The C. japonica cp genome is 131,810 bp in length, with 112 single copy genes and two duplicated (trnI-CAU, trnQ-UUG) genes that give a total of 116 genes. Compared to other land plant cp genomes, the C. japonica cp has lost one of the relevant large inverted repeats (IRs) found in angiosperms, fern, liverwort, and gymnosperms, such as Cycas and Gingko, and additionally has completely lost its trnR-CCG, partially lost its trnT-GGU, and shows diversification of accD. The genomic structure of the C. japonica cp genome also differs significantly from those of other plant species. For example, we estimate that a minimum of 15 inversions would be required to transform the gene organization of the Pinus thunbergii cp genome into that of C. japonica. In the C. japonica cp genome, direct repeat and inverted repeat sequences are observed at the inversion and translocation endpoints, and these sequences may be associated with the genomic rearrangements.
Conclusion: The observed differences in genomic structure between C. japonica and other land plants, including pines, strongly support the theory that the large IRs stabilize the cp genome. Furthermore, the deleted large IR and the numerous genomic rearrangements that have occurred in the C. japonica cp genome provide new insights into both the evolutionary lineage of coniferous species in gymnosperm and the evolution of the cp genome.
Figures
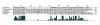







References
-
- Shinozaki K, Ohme M, Tanaka M, Wakasugi T, Hayashida N, Matsubayashi T, Zaita N, Chunwongse J, Obokata J, Yamaguchi-Shinozaki K, Ohto C, Torazawa K, Meng BY, Sugita M, Deno H, Kamogashira T, Yamada K, Kusuda J, Takaiwa F, Kato A, Tohdoh N, Shimada H, Sugiura M. The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 1986;5:2043–2049. - PMC - PubMed
-
- Ohyama K, Fukuzawa H, Kohchi T, Shirai H, Sano T, Sano S, Umesono K, Shiki Y, Takeuchi M, Chang Z, Aota S, Inokuchi H, Ozeki H. Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature. 1986;322:572–574. doi: 10.1038/322572a0. - DOI
-
- Jansen RK, Kaittanis C, Saski C, Lee SB, Tomkins J, Alverson AJ, Daniell H. Phylogenetic analysis of Vitis (Vitaceae) based on complete chloroplast genome sequences: effects of taxon sampling and phylogenetic methods on resolving relationships among rosids. BMC Evol Biol. 2006;6:32. doi: 10.1186/1471-2148-6-32. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

