"Nought may endure but mutability": spliceosome dynamics and the regulation of splicing
- PMID: 18570869
- PMCID: PMC2610350
- DOI: 10.1016/j.molcel.2008.04.013
"Nought may endure but mutability": spliceosome dynamics and the regulation of splicing
Abstract
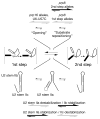
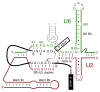
The spliceosome is both compositionally and conformationally dynamic. Each transition along the splicing pathway presents an opportunity for progression, pausing, or discard, allowing splice site choice to be regulated throughout both the assembly and catalytic phases of the reaction.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

