Aven-dependent activation of ATM following DNA damage
- PMID: 18571408
- PMCID: PMC2691717
- DOI: 10.1016/j.cub.2008.05.045
Aven-dependent activation of ATM following DNA damage
Abstract
Background: In response to DNA damage, cells undergo either cell-cycle arrest or apoptosis, depending on the extent of damage and the cell's capacity for DNA repair. Cell-cycle arrest induced by double-stranded DNA breaks depends on activation of the ataxia-telangiectasia (ATM) protein kinase, which phosphorylates cell-cycle effectors such as Chk2 and p53 to inhibit cell-cycle progression. ATM is recruited to double-stranded DNA breaks by a complex of sensor proteins, including Mre11/Rad50/Nbs1, resulting in autophosphorylation, monomerization, and activation of ATM kinase.
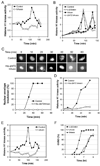
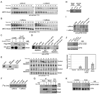
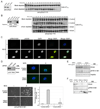
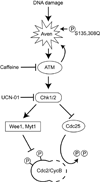
Results: In characterizing Aven protein, a previously reported apoptotic inhibitor, we have found that Aven can function as an ATM activator to inhibit G2/M progression. Aven bound to ATM and Aven overexpressed in cycling Xenopus egg extracts prevented mitotic entry and induced phosphorylation of ATM and its substrates. Immunodepletion of endogenous Aven allowed mitotic entry even in the presence of damaged DNA, and RNAi-mediated knockdown of Aven in human cells prevented autophosphorylation of ATM at an activating site (S1981) in response to DNA damage. Interestingly, Aven is also a substrate of the ATM kinase. Mutation of ATM-mediated phosphorylation sites on Aven reduced its ability to activate ATM, suggesting that Aven activation of ATM after DNA damage is enhanced by ATM-mediated Aven phosphorylation.
Conclusions: These results identify Aven as a new ATM activator and describe a positive feedback loop operating between Aven and ATM. In aggregate, these findings place Aven, a known apoptotic inhibitor, as a critical transducer of the DNA-damage signal.
Figures


References
-
- Lew DJ, Kornbluth S. Regulatory roles of cyclin dependent kinase phosphorylation in cell cycle control. Current opinion in cell biology. 1996;8:795–804. - PubMed
-
- Coleman TR, Dunphy WG. Cdc2 regulatory factors. Current opinion in cell biology. 1994;6:877–882. - PubMed
-
- Abraham RT. Cell cycle checkpoint signaling through the ATM and ATR kinases. Genes & development. 2001;15:2177–2196. - PubMed
-
- Raleigh JM, O'Connell MJ. The G(2) DNA damage checkpoint targets both Wee1 and Cdc25. Journal of cell science. 2000;113(Pt 10):1727–1736. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R01 NS034175/NS/NINDS NIH HHS/United States
- R01 CA102707/CA/NCI NIH HHS/United States
- R01 GM067225/GM/NIGMS NIH HHS/United States
- R01 GM043974/GM/NIGMS NIH HHS/United States
- R01 NS037402/NS/NINDS NIH HHS/United States
- R01 NS34704/NS/NINDS NIH HHS/United States
- R01 AI54952/AI/NIAID NIH HHS/United States
- R01 CA123250/CA/NCI NIH HHS/United States
- R01 GM070891/GM/NIGMS NIH HHS/United States
- R01 GM077875/GM/NIGMS NIH HHS/United States
- R01 AI054952/AI/NIAID NIH HHS/United States
- R0I GM67225/GM/NIGMS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

