Defective hepatitis B virus DNA is not associated with disease status but is reduced by polymerase mutations associated with drug resistance
- PMID: 18571815
- PMCID: PMC2669111
- DOI: 10.1002/hep.22386
Defective hepatitis B virus DNA is not associated with disease status but is reduced by polymerase mutations associated with drug resistance
Abstract
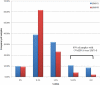
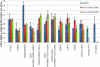
Defective hepatitis B virus DNA (dDNA) is reverse-transcribed from spliced hepatitis B virus (HBV) pregenomic messenger RNA (pgRNA) and has been identified in patients with chronic HBV (CH-B). The major 2.2-kb spliced pgRNA encodes a novel HBV gene product, the hepatitis B splice protein (HBSP) via a deletion and frame shift within the polymerase. Although spliced RNA and HBSP expression have been associated with increased HBV DNA levels and liver fibrosis, the role of dDNA in HBV-associated disease is largely undefined. Our aims were to (1) compare the relative proportions of dDNA (% dDNA) in a range of HBV-infected serum samples, including patients with human immunodeficiency virus (HIV)/HBV coinfection and HBV-monoinfected persons with differing severities of liver disease, and (2) determine the effect of mutations associated with drug resistance on defective DNA production. Defective DNA was detected in 90% of persons with CH-B. There was no significant difference in the relative abundance of dDNA between the monoinfected and HIV/HBV-coinfected groups. We also found no association between the % dDNA and alanine aminotransferase, hepatitis B e antigen status, HBV DNA levels, fibrosis levels, compensated or decompensated liver cirrhosis, genotype, or drug treatment. However, the % dDNA was significantly lower in individuals infected with lamivudine-resistant (LMV-R) HBV compared with wild-type HBV (P < 0.0001), indicating that antiviral drug resistance alters the balance between defective and genomic length DNA in circulation. Experiments in vitro using HBV encoding LMV-R mutations confirmed these results.
Conclusion: Our results identified no association between dDNA and parameters associated with disease status and suggested that the relative abundance of dDNA is largely dependent on the integrity of the HBV polymerase and is unrelated to the severity of liver disease.
Figures

Similar articles
-
Correlation of the occurrence of YMDD mutations with HBV genotypes, HBV-DNA levels, and HBeAg status in Chinese patients with chronic hepatitis B during lamivudine treatment.Hepatobiliary Pancreat Dis Int. 2012 Apr;11(2):172-6. doi: 10.1016/s1499-3872(12)60144-1. Hepatobiliary Pancreat Dis Int. 2012. PMID: 22484586
-
Large-scale survey of naturally occurring HBV polymerase mutations associated with anti-HBV drug resistance in untreated patients with chronic hepatitis B.J Viral Hepat. 2011 Jul;18(7):e212-6. doi: 10.1111/j.1365-2893.2011.01435.x. Epub 2011 Jan 21. J Viral Hepat. 2011. PMID: 21692935
-
Differences of YMDD mutational patterns, precore/core promoter mutations, serum HBV DNA levels in lamivudine-resistant hepatitis B genotypes B and C.J Viral Hepat. 2007 Nov;14(11):767-74. doi: 10.1111/j.1365-2893.2007.00869.x. J Viral Hepat. 2007. PMID: 17927612
-
Low risk of lamivudine-resistant HBV and hepatic flares in treated HIV-HBV-coinfected patients from Côte d'Ivoire.Antivir Ther. 2015;20(6):643-54. doi: 10.3851/IMP2959. Epub 2015 Apr 8. Antivir Ther. 2015. PMID: 25852125 Clinical Trial.
-
Spacer Domain in Hepatitis B Virus Polymerase: Plugging a Hole or Performing a Role?J Virol. 2022 May 11;96(9):e0005122. doi: 10.1128/jvi.00051-22. Epub 2022 Apr 12. J Virol. 2022. PMID: 35412348 Free PMC article. Review.
Cited by
-
Hepatitis B virus spliced variants are associated with an impaired response to interferon therapy.Sci Rep. 2015 Nov 20;5:16459. doi: 10.1038/srep16459. Sci Rep. 2015. PMID: 26585041 Free PMC article.
-
Clinical Implications of Hepatitis B Virus RNA and Covalently Closed Circular DNA in Monitoring Patients with Chronic Hepatitis B Today with a Gaze into the Future: The Field Is Unprepared for a Sterilizing Cure.Genes (Basel). 2018 Oct 5;9(10):483. doi: 10.3390/genes9100483. Genes (Basel). 2018. PMID: 30301171 Free PMC article. Review.
-
Hepatitis B splice-generated protein antibodies in Syrian chronic hepatitis B patients: incidence and significance.Hepat Mon. 2014 Apr 16;14(4):e13166. doi: 10.5812/hepatmon.13166. eCollection 2014 Apr. Hepat Mon. 2014. PMID: 24829585 Free PMC article.
-
Human hepatitis B virus production in avian cells is characterized by enhanced RNA splicing and the presence of capsids containing shortened genomes.PLoS One. 2012;7(5):e37248. doi: 10.1371/journal.pone.0037248. Epub 2012 May 18. PLoS One. 2012. PMID: 22624002 Free PMC article.
-
Secreted hepatitis B virus splice variants differ by HBV genotype and across phases of chronic hepatitis B infection.J Viral Hepat. 2022 Aug;29(8):604-615. doi: 10.1111/jvh.13702. Epub 2022 May 28. J Viral Hepat. 2022. PMID: 35582878 Free PMC article.
References
-
- Guidotti LG, Rochford R, Chung J, Shapiro M, Purcell R, Chisari FV. Viral clearance without destruction of infected cells during acute HBV infection. Science. 1999;284:825–829. - PubMed
-
- Thio CL, Seaberg EC, Skolasky R, Jr, Phair J, Visscher B, Munoz A, et al. HIV-1, hepatitis B virus, and risk of liver-related mortality in the Multicenter Cohort Study (MACS) Lancet. 2002;360:1921–1926. - PubMed
-
- Bica I, McGovern B, Dhar R, Stone D, McGowan K, Scheib R, et al. Increasing mortality due to end-stage liver disease in patients with human immunodeficiency virus infection. Clin Infect Dis. 2001;32:492–497. - PubMed
-
- Revill PA, Littlejohn M, Ayres A, Yuen L, Colledge D, Bartholomeusz A, et al. Identification of a novel hepatitis B virus precore/core deletion mutant in HIV/hepatitis B virus co-infected individuals. AIDS. 2007;21:1701–1710. - PubMed
-
- Gunther S, Sommer G, Iwanska A, Will H. Heterogeneity and common features of defective hepatitis B virus genomes derived from spliced pregenomic RNA. Virology. 1997;238:363–371. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
