RNA folding: conformational statistics, folding kinetics, and ion electrostatics
- PMID: 18573079
- PMCID: PMC2473866
- DOI: 10.1146/annurev.biophys.37.032807.125957
RNA folding: conformational statistics, folding kinetics, and ion electrostatics
Abstract
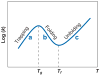
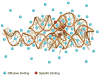
RNA folding is a remarkably complex problem that involves ion-mediated electrostatic interaction, conformational entropy, base pairing and stacking, and noncanonical interactions. During the past decade, results from a variety of experimental and theoretical studies pointed to (a) the potential ion correlation effect in Mg2+-RNA interactions, (b) the rugged energy landscapes and multistate RNA folding kinetics even for small RNA systems such as hairpins and pseudoknots, (c) the intraloop interactions and sequence-dependent loop free energy, and (d) the strong nonadditivity of chain entropy in RNA pseudoknot and other tertiary folds. Several related issues, which have not been thoroughly resolved, require combined approaches with thermodynamic and kinetic experiments, statistical mechanical modeling, and all-atom computer simulations.
Figures

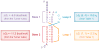
References
-
- Andresen K, Das R, Park HY, Smith H, Kwok LW, et al. Spatial distribution of competing ions around DNA in solution. Phys Rev Lett. 2004;93:248103. - PubMed
-
- Andronescu M, Zhang ZC, Condon A. Secondary structure prediction of interacting RNA molecules. J Mol Biol. 2005;345:987–1001. - PubMed
-
- Bai Y, Das R, Millett IS, Herschlag D, Doniach S. Probing counterion modulated repulsion and attraction between nucleic acid duplexes in solution. Proc Natl Acad Sci USA. 2005;102:1035–40. Experiment demonstrates that the Mg2+-induced force is not sufficient to cause a collapsed state for two short DNA helices tethered by a neutral short loop. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

