Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts
- PMID: 18573878
- PMCID: PMC2519735
- DOI: 10.1128/MCB.02056-07
Molecular identification and function of cis- and trans-acting determinants for petA transcript stability in Chlamydomonas reinhardtii chloroplasts
Abstract
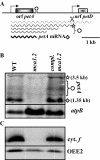
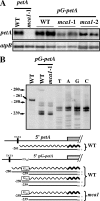
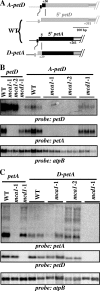
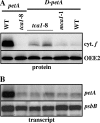
In organelles, the posttranscriptional steps of gene expression are tightly controlled by nucleus-encoded factors, most often acting in a gene-specific manner. Despite the molecular identification of a growing number of factors, their mode of action remains largely unknown. In the green alga Chlamydomonas reinhardtii, expression of the chloroplast petA gene, which codes for cytochrome f, depends on two specific nucleus-encoded factors. MCA1 controls the accumulation of the transcript, while TCA1 is required for its translation. We report here the cloning of MCA1, the first pentatricopeptide repeat protein functionally identified in this organism. By chloroplast transformation with modified petA genes, we investigated the function of MCA1 in vivo. We demonstrate that MCA1 acts on the very first 21 nucleotides of the petA 5' untranslated region to protect the whole transcript from 5'-->3' degradation but does not process the 5' end of the petA mRNA. MCA1 and TCA1 recognize adjacent targets and probably interact together for efficient expression of petA mRNA. MCA1, although not strictly required for translation, shows features of a translational enhancer, presumably by assisting the binding of TCA1 to its own target. Conversely, TCA1 participates to the full stabilization of the transcript through its interaction with MCA1.
Figures





References
-
- Andres, C., C. Lurin, and I. D. Small. 2007. The multifarious roles of PPR proteins in plant mitochondrial gene expression. Physiol. Plant 12914-22.
-
- Barkan, A., and M. Goldschmidt-Clermont. 2000. Participation of nuclear gene in chloroplast gene expression. Biochimie 82559-572. - PubMed
-
- Bollenbach, T. J., G. Schuster, and D. B. Stern. 2004. Cooperation of endo- and exoribonucleases in chloroplast mRNA turnover. Prog. Nucleic Acids Res. Mol. Biol. 78305-337. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
