The resistance of BALB/cJ mice to Yersinia pestis maps to the major histocompatibility complex of chromosome 17
- PMID: 18573896
- PMCID: PMC2519398
- DOI: 10.1128/IAI.00488-08
The resistance of BALB/cJ mice to Yersinia pestis maps to the major histocompatibility complex of chromosome 17
Abstract
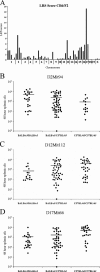
Yersinia pestis, the causative agent of plague, has been well studied at the molecular and genetic levels, but little is known about the role that host genes play in combating this highly lethal pathogen. We challenged several inbred strains of mice with Y. pestis and found that BALB/cJ mice are highly resistant compared to susceptible strains such as C57BL/6J. This resistance was observed only in BALB/cJ mice and not in other BALB/c substrains. Compared to C57BL/6J mice, the BALB/cJ strain exhibited reduced bacterial burden in the spleen and liver early after infection as well as lower levels of serum interleukin-6. These differences were evident 24 h postinfection and became more pronounced with time. Although a significant influx of neutrophils in the spleen and liver was exhibited in both strains, occlusive fibrinous thrombi resulting in necrosis of the surrounding tissue was observed only in C57BL/6J mice. In an effort to identify the gene(s) responsible for resistance, we measured total splenic bacteria in 95 F(2) mice 48 h postinfection and performed quantitative trait locus mapping using 58 microsatellite markers spaced throughout the genome. This analysis revealed a single nonrecessive plague resistance locus, designated prl1 (plague resistance locus 1), which coincides with the major histocompatibility complex of chromosome 17. A second screen of 95 backcrossed mice verified that this locus confers resistance to Y. pestis early in infection. Finally, eighth generation backcrossed mice harboring prl1 were found to maintain resistance in the susceptible C57BL/6J background. These results identify a novel genetic locus in BALB/cJ mice that confers resistance to Y. pestis.
Figures



Similar articles
-
Substrains of 129 mice are resistant to Yersinia pestis KIM5: implications for interleukin-10-deficient mice.Infect Immun. 2009 Jan;77(1):367-73. doi: 10.1128/IAI.01057-08. Epub 2008 Oct 27. Infect Immun. 2009. PMID: 18955473 Free PMC article.
-
Delayed inflammatory response to primary pneumonic plague occurs in both outbred and inbred mice.Infect Immun. 2007 Feb;75(2):697-705. doi: 10.1128/IAI.00403-06. Epub 2006 Nov 13. Infect Immun. 2007. PMID: 17101642 Free PMC article.
-
The plague virulence protein YopM targets the innate immune response by causing a global depletion of NK cells.Infect Immun. 2004 Aug;72(8):4589-602. doi: 10.1128/IAI.72.8.4589-4602.2004. Infect Immun. 2004. PMID: 15271919 Free PMC article.
-
Immunology of Yersinia pestis Infection.Adv Exp Med Biol. 2016;918:273-292. doi: 10.1007/978-94-024-0890-4_10. Adv Exp Med Biol. 2016. PMID: 27722867 Review.
-
Role of immune response in Yersinia pestis infection.J Infect Dev Ctries. 2011 Sep 14;5(9):628-39. doi: 10.3855/jidc.1999. J Infect Dev Ctries. 2011. PMID: 21918303 Review.
Cited by
-
The dependence of the Yersinia pestis capsule on pathogenesis is influenced by the mouse background.Infect Immun. 2011 Feb;79(2):644-52. doi: 10.1128/IAI.00981-10. Epub 2010 Nov 29. Infect Immun. 2011. PMID: 21115720 Free PMC article.
-
Substrains of 129 mice are resistant to Yersinia pestis KIM5: implications for interleukin-10-deficient mice.Infect Immun. 2009 Jan;77(1):367-73. doi: 10.1128/IAI.01057-08. Epub 2008 Oct 27. Infect Immun. 2009. PMID: 18955473 Free PMC article.
-
Resistance of Mice of the 129 Background to Yersinia pestis Maps to Multiple Loci on Chromosome 1.Infect Immun. 2016 Sep 19;84(10):2904-13. doi: 10.1128/IAI.00488-16. Print 2016 Oct. Infect Immun. 2016. PMID: 27481241 Free PMC article.
-
Interleukin-10 induction is an important virulence function of the Yersinia pseudotuberculosis type III effector YopM.Infect Immun. 2012 Jul;80(7):2519-27. doi: 10.1128/IAI.06364-11. Epub 2012 Apr 30. Infect Immun. 2012. PMID: 22547545 Free PMC article.
-
Functionally distinct regions of the locus Leishmania major response 15 control IgE or IFNγ level in addition to skin lesions.Front Immunol. 2023 Aug 3;14:1145269. doi: 10.3389/fimmu.2023.1145269. eCollection 2023. Front Immunol. 2023. PMID: 37600780 Free PMC article.
References
-
- Autenrieth, I. B., and J. Heesemann. 1992. In vivo neutralization of tumor necrosis factor-alpha and interferon-gamma abrogates resistance to Yersinia enterocolitica infection in mice. Med. Microbiol. Immunol. 181333-338. - PubMed
-
- Bohn, E., and I. B. Autenrieth. 1996. IL-12 is essential for resistance against Yersinia enterocolitica by triggering IFN-gamma production in NK cells and CD4+ T cells. J. Immunol. 1561458-1468. - PubMed
-
- Cornelis, G. R. 2002. The Yersinia Ysc-Yop “type III” weaponry. Nat. Rev. Mol. Cell Biol. 3742-752. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

